A genome-scale metabolic model of a globally disseminated hyperinvasive M1 strain of Streptococcus pyogenes
- PMID: 39158303
- PMCID: PMC11406949
- DOI: 10.1128/msystems.00736-24
A genome-scale metabolic model of a globally disseminated hyperinvasive M1 strain of Streptococcus pyogenes
Abstract
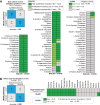
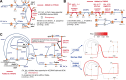
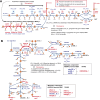
Streptococcus pyogenes is responsible for a range of diseases in humans contributing significantly to morbidity and mortality. Among more than 200 serotypes of S. pyogenes, serotype M1 strains hold the greatest clinical relevance due to their high prevalence in severe human infections. To enhance our understanding of pathogenesis and discovery of potential therapeutic approaches, we have developed the first genome-scale metabolic model (GEM) for a serotype M1 S. pyogenes strain, which we name iYH543. The curation of iYH543 involved cross-referencing a draft GEM of S. pyogenes serotype M1 from the AGORA2 database with gene essentiality and autotrophy data obtained from transposon mutagenesis-based and growth screens. We achieved a 92.6% (503/543 genes) accuracy in predicting gene essentiality and a 95% (19/20 amino acids) accuracy in predicting amino acid auxotrophy. Additionally, Biolog Phenotype microarrays were employed to examine the growth phenotypes of S. pyogenes, which further contributed to the refinement of iYH543. Notably, iYH543 demonstrated 88% accuracy (168/190 carbon sources) in predicting growth on various sole carbon sources. Discrepancies observed between iYH543 and the actual behavior of living S. pyogenes highlighted areas of uncertainty in the current understanding of S. pyogenes metabolism. iYH543 offers novel insights and hypotheses that can guide future research efforts and ultimately inform novel therapeutic strategies.IMPORTANCEGenome-scale models (GEMs) play a crucial role in investigating bacterial metabolism, predicting the effects of inhibiting specific metabolic genes and pathways, and aiding in the identification of potential drug targets. Here, we have developed the first GEM for the S. pyogenes highly virulent serotype, M1, which we name iYH543. The iYH543 achieved high accuracy in predicting gene essentiality. We also show that the knowledge obtained by substituting actual measurement values for iYH543 helps us gain insights that connect metabolism and virulence. iYH543 will serve as a useful tool for rational drug design targeting S. pyogenes metabolism and computational screening to investigate the interplay between inhibiting virulence factor synthesis and growth.
Keywords: Streptococcus pyogenes; auxotrophy; carbon sources; essential gene; genome-scale model; metabolic modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Grants and funding
- R37 AI052453/AI/NIAID NIH HHS/United States
- R01-AI173689/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- K99 DE029228/DE/NIDCR NIH HHS/United States
- 22K09924,22H03262,Overseas Research Fellowship/MEXT | Japan Society for the Promotion of Science (JSPS)
- R01 AI173689/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
