The Hemagglutinin of Influenza A Virus Induces Ferroptosis to Facilitate Viral Replication
- PMID: 39159143
- PMCID: PMC11497066
- DOI: 10.1002/advs.202404365
The Hemagglutinin of Influenza A Virus Induces Ferroptosis to Facilitate Viral Replication
Abstract
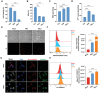
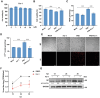
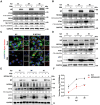
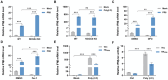
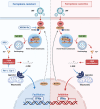
Ferroptosis is a novel form of cell death caused by the accumulation of lipid peroxides in an iron-dependent manner. However, the precise mechanism underlying the exploitation of ferroptosis by influenza A viruses (IAV) remains unclear. The results demonstrate that IAV promotes its own replication through ferritinophagy by sensitizing cells to ferroptosis, with hemagglutinin identified as a key trigger in this process. Hemagglutinin interacts with autophagic receptors nuclear receptor coactivator 4 (NCOA4) and tax1-binding protein 1 (TAX1BP1), facilitating the formation of ferritin-NCOA4 condensates and inducing ferritinophagy. Further investigation shows that hemagglutinin-induced ferritinophagy causes cellular lipid peroxidation, inhibits aggregation of mitochondrial antiviral signaling protein (MAVS), and suppresses the type I interferon response, thereby contributing to viral replication. Collectively, a novel mechanism by which IAV hemagglutinin induces ferritinophagy resulting in cellular lipid peroxidation, consequently impairing MAVS-mediated antiviral immunity, is revealed.
Keywords: autophagy; ferroptosis; influenza a virus; innate immunity; mitochondrial antiviral signaling protein (MAVS).
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
