Predicting Antigen-Specificities of Orphan T Cell Receptors from Cancer Patients with TCRpcDist
- PMID: 39159239
- PMCID: PMC11516110
- DOI: 10.1002/advs.202405949
Predicting Antigen-Specificities of Orphan T Cell Receptors from Cancer Patients with TCRpcDist
Abstract
Approaches to analyze and cluster T-cell receptor (TCR) repertoires to reflect antigen specificity are critical for the diagnosis and prognosis of immune-related diseases and the development of personalized therapies. Sequence-based approaches showed success but remain restrictive, especially when the amount of experimental data used for the training is scarce. Structure-based approaches which represent powerful alternatives, notably to optimize TCRs affinity toward specific epitopes, show limitations for large-scale predictions. To handle these challenges, TCRpcDist is presented, a 3D-based approach that calculates similarities between TCRs using a metric related to the physico-chemical properties of the loop residues predicted to interact with the epitope. By exploiting private and public datasets and comparing TCRpcDist with competing approaches, it is demonstrated that TCRpcDist can accurately identify groups of TCRs that are likely to bind the same epitopes. Importantly, the ability of TCRpcDist is experimentally validated to determine antigen specificities (neoantigens and tumor-associated antigens) of orphan tumor-infiltrating lymphocytes (TILs) in cancer patients. TCRpcDist is thus a promising approach to support TCR repertoire analysis and TCR deorphanization for individualized treatments including cancer immunotherapies.
Keywords: deorphanization; epitope specificity; specificity prediction; t cell receptors (TCRs); tcr clustering; tumor antigens.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
VZ is consultant for Cellestia Biotech. GC has received grants, research support or is coinvestigator in clinical trials by Bristol‐Myers‐Squibb, Celgene, Boehringer Ingelheim, Tigen, Roche, Iovance and Kite. GC has received honoraria for consultations or presentations by Roche, Genentech, BMS, AstraZeneca, Sanofi‐Aventis, Nextcure and GeneosTx. GC has patents in the domain of antibodies and vaccines targeting the tumor vasculature as well as technologies related to T‐cell expansion and engineering for T‐cell therapy. GC receives royalties from the University of Pennsylvania. SB and AH have patents in technologies related to T‐cell expansion and engineering for T‐cell therapy. Other authors declare no competing interests.
Figures
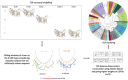

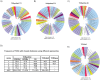

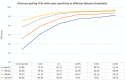


References
-
- a) Borrman T., Cimons J., Cosiano M., Purcaro M., Pierce B. G., Baker B. M., Weng Z., Proteins 2017, 85, 908; - PMC - PubMed
- b) Gálvez J., Gálvez J. J., García‐Peñarrubia P., Front. Immunol. 2019, 10, 349; - PMC - PubMed
- c) Schmidt J., Chiffelle J., Perez M. A. S., Magnin M., Bobisse S., Arnaud M., Genolet R., Cesbron J., Barras D., Navarro Rodrigo B., Benedetti F., Michel A., Queiroz L., Baumgaertner P., Guillaume P., Hebeisen M., Michielin O., Nguyen‐Ngoc T., Huber F., Irving M., Tissot‐Renaud S., Stevenson B. J., Rusakiewicz S., Dangaj Laniti D., Bassani‐Sternberg M., Rufer N., Gfeller D., Kandalaft L. E., Speiser D. E., Zoete V., et al., Nat. Commun. 2023, 14, 3188. - PMC - PubMed
-
- Chiffelle J., Genolet R., Perez M. A., Coukos G., Zoete V., Harari A., Curr. Opin. Biotechnol. 2020, 65, 284. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
