Iron-sulfur cluster loss in mitochondrial CISD1 mediates PINK1 loss-of-function phenotypes
- PMID: 39159312
- PMCID: PMC11383524
- DOI: 10.7554/eLife.97027
Iron-sulfur cluster loss in mitochondrial CISD1 mediates PINK1 loss-of-function phenotypes
Abstract
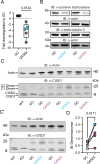
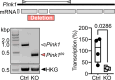
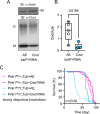
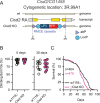
Parkinson's disease (PD) is characterized by the progressive loss of dopaminergic neurons in the substantia nigra of the midbrain. Familial cases of PD are often caused by mutations of PTEN-induced kinase 1 (PINK1) and the ubiquitin ligase Parkin, both pivotal in maintaining mitochondrial quality control. CISD1, a homodimeric mitochondrial iron-sulfur-binding protein, is a major target of Parkin-mediated ubiquitination. We here discovered a heightened propensity of CISD1 to form dimers in Pink1 mutant flies and in dopaminergic neurons from PINK1 mutation patients. The dimer consists of two monomers that are covalently linked by a disulfide bridge. In this conformation CISD1 cannot coordinate the iron-sulfur cofactor. Overexpressing Cisd, the Drosophila ortholog of CISD1, and a mutant Cisd incapable of binding the iron-sulfur cluster in Drosophila reduced climbing ability and lifespan. This was more pronounced with mutant Cisd and aggravated in Pink1 mutant flies. Complete loss of Cisd, in contrast, rescued all detrimental effects of Pink1 mutation on climbing ability, wing posture, dopamine levels, lifespan, and mitochondrial ultrastructure. Our results suggest that Cisd, probably iron-depleted Cisd, operates downstream of Pink1 shedding light on PD pathophysiology and implicating CISD1 as a potential therapeutic target.
Keywords: D. melanogaster; Drosophila; Parkinson's disease; cell biology; dopaminergic neurons; human; iron; mitochondria; mouse; neuroscience; oxidative distress.
Plain language summary
Parkinson’s disease affects millions of people worldwide, causing progressively worse symptoms like stiffness, tremors and difficulty moving. These issues result from the death of neurons in the brain that produce the neurotransmitter dopamine. While most cases have no known cause, 10 to 15 per cent are due to inherited gene mutations. This includes mutations in the genes that code for the proteins PINK1 and Parkin which are essential for maintaining healthy mitochondria, the powerhouse of the cell. Mutations in this quality control system affect a protein called CISD1, which sits within the outer surface of the mitochondria. CISD1 contains a cluster of iron and sulfur ions, and is involved in regulating iron levels and mitochondrial energy production. However, its role in inherited cases of Parkinson’s disease, particularly those related to mutations in PINK1 and Parkin, is poorly understood. To understand the impact of CISD1, Bitar et al. studied genetically modified fruit flies and dopamine-producing neurons from Parkinson’s patients with PINK1 mutations. This revealed that losing PINK1 activity led to higher levels of CISD1 proteins which lacked the iron-sulfur cluster due to a bond forming between two CISD1 molecules. Reducing levels of the CISD1-equivalent protein in the flies helped to alleviate most of the symptoms caused by PINK1 and Parkin gene mutations, such as difficulties climbing and impaired wing posture. These findings suggest that iron-depleted CISD1 contributes to the symptoms associated with Parkinson’s disease, underscoring its potential as a drug target. Drugs that target CISD1 already exist, which could ease the way for further research. Recent studies have shown that cases of Parkinson’s related to mutations in PINK-1 share features with some non-inherited instances of the disease, suggesting that this approach could potentially benefit many patients.
© 2024, Bitar et al.
Conflict of interest statement
SB, TB, CW, MA, LR, PZ, TS, IR, VV, BM, EH, GA, RK, LZ, AM No competing interests declared
Figures






Update of
References
-
- Antico O, Ordureau A, Stevens M, Singh F, Nirujogi RS, Gierlinski M, Barini E, Rickwood ML, Prescott A, Toth R, Ganley IG, Harper JW, Muqit MMK. Global ubiquitylation analysis of mitochondria in primary neurons identifies endogenous Parkin targets following activation of PINK1. Science Advances. 2021;7:eabj0722. doi: 10.1126/sciadv.abj0722. - DOI - PMC - PubMed
-
- Bus C, Zizmare L, Feldkaemper M, Geisler S, Zarani M, Schaedler A, Klose F, Admard J, Mageean CJ, Arena G, Fallier-Becker P, Ugun-Klusek A, Maruszczak KK, Kapolou K, Schmid B, Rapaport D, Ueffing M, Casadei N, Krüger R, Gasser T, Vogt Weisenhorn DM, Kahle PJ, Trautwein C, Gloeckner CJ, Fitzgerald JC. human dopaminergic neurons lacking PINK1 exhibit disrupted dopamine metabolism related to vitamin B6 co-factors. iScience. 2020;23:101797. doi: 10.1016/j.isci.2020.101797. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

