The involvement of the Stat1/Nrf2 pathway in exacerbating Crizotinib-induced liver injury: implications for ferroptosis
- PMID: 39160159
- PMCID: PMC11333746
- DOI: 10.1038/s41419-024-06993-z
The involvement of the Stat1/Nrf2 pathway in exacerbating Crizotinib-induced liver injury: implications for ferroptosis
Abstract
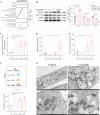
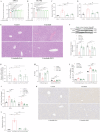
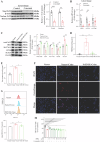
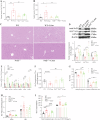
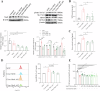
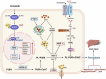
Crizotinib carries an FDA hepatotoxicity warning, yet analysis of the FAERS database suggests that the severity of its hepatotoxicity risks, including progression to hepatitis and liver failure, might be underreported. However, the underlying mechanism remains poorly understood, and effective intervention strategies are lacking. Here, mRNA-sequencing analysis, along with KEGG and GO analyses, revealed that DEGs linked to Crizotinib-induced hepatotoxicity predominantly associate with the ferroptosis pathway which was identified as the principal mechanism behind Crizotinib-induced hepatocyte death. Furthermore, we found that ferroptosis inhibitors, namely Ferrostatin-1 and Deferoxamine mesylate, significantly reduced Crizotinib-induced hepatotoxicity and ferroptosis in both in vivo and in vitro settings. We have also discovered that overexpression of AAV8-mediated Nrf2 could mitigate Crizotinib-induced hepatotoxicity and ferroptosis in vivo by restoring the imbalance in glutathione metabolism, iron homeostasis, and lipid peroxidation. Additionally, both Stat1 deficiency and the Stat1 inhibitor NSC118218 were found to reduce Crizotinib-induced ferroptosis. Mechanistically, Crizotinib induces the phosphorylation of Stat1 at Ser727 but not Tyr701, promoting the transcriptional inhibition of Nrf2 expression after its entry into the nucleus to promote ferroptosis. Meanwhile, we found that MgIG and GA protected against hepatotoxicity to counteract ferroptosis without affecting or compromising the anti-cancer activity of Crizotinib, with a mechanism potentially related to the Stat1/Nrf2 pathway. Overall, our findings identify that the phosphorylation activation of Stat1 Ser727, rather than Tyr701, promotes ferroptosis through transcriptional inhibition of Nrf2, and highlight MgIG and GA as potential therapeutic approaches to enhance the safety of Crizotinib-based cancer therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Blackhall F, Ross Camidge D, Shaw AT, Soria JC, Solomon BJ, Mok T, et al. Final results of the large-scale multinational trial PROFILE 1005: efficacy and safety of crizotinib in previously treated patients with advanced/metastatic ALK-positive non-small-cell lung cancer. ESMO Open. 2017;2:e000219. 10.1136/esmoopen-2017-000219 - DOI - PMC - PubMed
-
- Zhu Y, Jiang ZZ, Zhang LY. Research progress of hepatocyte death mode and therapeutic drugs in drug-induced liver injury. Drug Eval Study. 2021;44:97–1104.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

