The integrated genomic surveillance system of Andalusia (SIEGA) provides a One Health regional resource connected with the clinic
- PMID: 39160186
- PMCID: PMC11333592
- DOI: 10.1038/s41598-024-70107-0
The integrated genomic surveillance system of Andalusia (SIEGA) provides a One Health regional resource connected with the clinic
Abstract
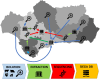
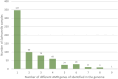
The One Health approach, recognizing the interconnectedness of human, animal, and environmental health, has gained significance amid emerging zoonotic diseases and antibiotic resistance concerns. This paper aims to demonstrate the utility of a collaborative tool, the SIEGA, for monitoring infectious diseases across domains, fostering a comprehensive understanding of disease dynamics and risk factors, highlighting the pivotal role of One Health surveillance systems. Raw whole-genome sequencing is processed through different species-specific open software that additionally reports the presence of genes associated to anti-microbial resistances and virulence. The SIEGA application is a Laboratory Information Management System, that allows customizing reports, detect transmission chains, and promptly alert on alarming genetic similarities. The SIEGA initiative has successfully accumulated a comprehensive collection of more than 1900 bacterial genomes, including Salmonella enterica, Listeria monocytogenes, Campylobacter jejuni, Escherichia coli, Yersinia enterocolitica and Legionella pneumophila, showcasing its potential in monitoring pathogen transmission, resistance patterns, and virulence factors. SIEGA enables customizable reports and prompt detection of transmission chains, highlighting its contribution to enhancing vigilance and response capabilities. Here we show the potential of genomics in One Health surveillance when supported by an appropriate bioinformatic tool. By facilitating precise disease control strategies and antimicrobial resistance management, SIEGA enhances global health security and reduces the burden of infectious diseases. The integration of health data from humans, animals, and the environment, coupled with advanced genomics, underscores the importance of a holistic One Health approach in mitigating health threats.
Keywords: AMR; Epidemiology; One health; Resistances; Surveillance; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Queenan, K. et al. Roadmap to a One Health agenda 2030. CABI Rev.10.1079/PAVSNNR201712014 (2017). 10.1079/PAVSNNR201712014 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

