A broad-spectrum vaccine candidate against H5 viruses bearing different sub-clade 2.3.4.4 HA genes
- PMID: 39160189
- PMCID: PMC11333769
- DOI: 10.1038/s41541-024-00947-4
A broad-spectrum vaccine candidate against H5 viruses bearing different sub-clade 2.3.4.4 HA genes
Abstract
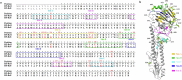
The global spread of H5 clade 2.3.4.4 highly pathogenic avian influenza (HPAI) viruses threatens poultry and public health. The continuous circulation of these viruses has led to their considerable genetic and antigenic evolution, resulting in the formation of eight subclades (2.3.4.4a-h). Here, we examined the antigenic sites that determine the antigenic differences between two H5 vaccine strains, H5-Re8 (clade 2.3.4.4g) and H5-Re11 (clade 2.3.4.4h). Epitope mapping data revealed that all eight identified antigenic sites were located within two classical antigenic regions, with five sites in region A (positions 115, 120, 124, 126, and 140) and three in region B (positions 151, 156, and 185). Through antigenic cartography analysis of mutants with varying numbers of substitutions, we confirmed that a combination of mutations in these eight sites reverses the antigenicity of H5-Re11 to that of H5-Re8, and vice versa. More importantly, our analyses identified H5-Re11_Q115L/R120S/A156T (H5-Re11 + 3) as a promising candidate for a broad-spectrum vaccine, positioned centrally in the antigenic map, and offering potential universal protection against all variants within the clade 2.3.4.4. H5-Re11 + 3 serum has better cross-reactivity than sera generated with other 2.3.4.4 vaccines, and H5-Re11 + 3 vaccine provided 100% protection of chickens against antigenically drifted H5 viruses from various 2.3.4.4 antigenic groups. Our findings suggest that antigenic regions A and B are immunodominant in H5 viruses, and that antigenic cartography-guided vaccine design is a promising strategy for selecting a broad-spectrum vaccine.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Yoon, S. W., Webby, R. J. & Webster, R. G. Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol.385, 359–375 (2014). - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

