Cancer tissue of origin constrains the growth and metabolism of metastases
- PMID: 39160333
- PMCID: PMC11450831
- DOI: 10.1038/s42255-024-01105-9
Cancer tissue of origin constrains the growth and metabolism of metastases
Abstract
Metastases arise from subsets of cancer cells that disseminate from the primary tumour1,2. The ability of cancer cells to thrive in a new tissue site is influenced by genetic and epigenetic changes that are important for disease initiation and progression, but these factors alone do not predict if and where cancers metastasize3,4. Specific cancer types metastasize to consistent subsets of tissues, suggesting that primary tumour-associated factors influence where cancers can grow. We find primary and metastatic pancreatic tumours have metabolic similarities and that the tumour-initiating capacity and proliferation of both primary-derived and metastasis-derived cells is favoured in the primary site relative to the metastatic site. Moreover, propagating cells as tumours in the lung or the liver does not enhance their relative ability to form large tumours in those sites, change their preference to grow in the primary site, nor stably alter aspects of their metabolism relative to primary tumours. Primary liver and lung cancer cells also exhibit a preference to grow in their primary site relative to metastatic sites. These data suggest cancer tissue of origin influences both primary and metastatic tumour metabolism and may impact where cancer cells can metastasize.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures





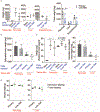


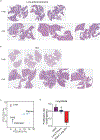





References
-
- Yang D. et al. Lineage Recording Reveals the Phylodynamics, Plasticity and Paths of Tumor Evolution. bioRxiv 13, 2021.10.12.464111 (2021).
MeSH terms
Grants and funding
- K99 CA234221/CA/NCI NIH HHS/United States
- U01 CA210171/CA/NCI NIH HHS/United States
- R01 CA259253/CA/NCI NIH HHS/United States
- R35 GM131884/GM/NIGMS NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32 GM144273/GM/NIGMS NIH HHS/United States
- F31 CA271787/CA/NCI NIH HHS/United States
- P30CA14051/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- F30 HL156404/HL/NHLBI NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- R01 CA201276/CA/NCI NIH HHS/United States
- R35 CA242379/CA/NCI NIH HHS/United States
- R01CA201276/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R35CA242379/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources

