Genomic characterization of AML with aberrations of chromosome 7: a multinational cohort of 519 patients
- PMID: 39160538
- PMCID: PMC11331663
- DOI: 10.1186/s13045-024-01590-1
Genomic characterization of AML with aberrations of chromosome 7: a multinational cohort of 519 patients
Abstract
Background: Deletions and partial losses of chromosome 7 (chr7) are frequent in acute myeloid leukemia (AML) and are linked to dismal outcome. However, the genomic landscape and prognostic impact of concomitant genetic aberrations remain incompletely understood.
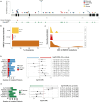
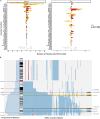
Methods: To discover genetic lesions in adult AML patients with aberrations of chromosome 7 [abn(7)], 60 paired diagnostic/remission samples were investigated by whole-exome sequencing in the exploration cohort. Subsequently, a gene panel including 66 genes and a SNP backbone for copy-number variation detection was designed and applied to the remaining samples of the validation cohort. In total, 519 patients were investigated, of which 415 received intensive induction treatment, typically containing a combination of cytarabine and anthracyclines.
Results: In the exploration cohort, the most frequently mutated gene was TP53 (33%), followed by epigenetic regulators (DNMT3A, KMT2C, IDH2) and signaling genes (NRAS, PTPN11). Thirty percent of 519 patients harbored ≥ 1 mutation in genes located in commonly deleted regions of chr7-most frequently affecting KMT2C (16%) and EZH2 (10%). KMT2C mutations were often subclonal and enriched in patients with del(7q), de novo or core-binding factor AML (45%). Cancer cell fraction analysis and reconstruction of mutation acquisition identified TP53 mutations as mainly disease-initiating events, while del(7q) or -7 appeared as subclonal events in one-third of cases. Multivariable analysis identified five genetic lesions with significant prognostic impact in intensively treated AML patients with abn(7). Mutations in TP53 and PTPN11 (11%) showed the strongest association with worse overall survival (OS, TP53: hazard ratio [HR], 2.53 [95% CI 1.66-3.86]; P < 0.001; PTPN11: HR, 2.24 [95% CI 1.56-3.22]; P < 0.001) and relapse-free survival (RFS, TP53: HR, 2.3 [95% CI 1.25-4.26]; P = 0.008; PTPN11: HR, 2.32 [95% CI 1.33-4.04]; P = 0.003). By contrast, IDH2-mutated patients (9%) displayed prolonged OS (HR, 0.51 [95% CI 0.30-0.88]; P = 0.0015) and durable responses (RFS: HR, 0.5 [95% CI 0.26-0.96]; P = 0.036).
Conclusion: This work unraveled formerly underestimated genetic lesions and provides a comprehensive overview of the spectrum of recurrent gene mutations and their clinical relevance in AML with abn(7). KMT2C mutations are among the most frequent gene mutations in this heterogeneous AML subgroup and warrant further functional investigation.
Keywords: IDH2; KMT2C; PTPN11; TP53; AML; Complex karyotype; Monosomy 7; del(7q).
© 2024. The Author(s).
Conflict of interest statement
F.D. reports personal fees from AbbVie, Amgen, Astra Zeneca, BeiGene, Gilead, Incyte, Novartis, and Roche outside the submitted work. L.B. reports advisory roles for Abbvie, Amgen, Astellas, Bristol-Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Pfizer, Sanofi, Servier; as well as research funding from Bayer, Jazz Pharmaceuticals. A.H. reports travel fees outside of the submitted work from Jazz Pharmaceuticals. M.T. and P.S. declare no COI related to this work. E.J. reports current employment at AstraZeneca. H.D. declares consultancy with honoraria from AbbVie, AstraZeneca, Gilead, Janssen, Jazz, Pfizer, Servier, Stemline, Syndax and Clinical Research funding to the Institution from AbbVie, Astellas, Bristol Myers Squibb, Celgene, Jazz Pharmaceuticals, Kronos Bio and Servier. J.E. reports participation in Advisory Boards of Abbvie, Novartis, Astellas, Jazz Pharmaceuticals, BMS-Celgene, Pfizer, Amgen and Research grants from Novartis and Jazz Pharmaceuticals. J.K. reports personal fees from BMS/Celgene, Takeda, Janssen, Abbvie, Sanofi, Pfizer and Jazz Pharmaceuticals outside the submitted work. M.H. reports research funding to institution from Abbvie, Servier, Astellas, BergenBio, Glycostem, Jazz Pharmaceuticals, Karyopharm, Loxo Oncology, Novartis, PinotBio, honoraria by Abbvie, Bristol Myers Squibb, Janssen, Jazz Pharmaceuticals, Pfizer, Qiagen, Servier, Sobi, and consultancy fees by AvenCell, Abbvie, Astellas, Glycostem, Janssen, LabDelbert, Miltenyi, Novartis, Pfizer, PinotBio and Servier.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

