Genome-wide identification of HSP90 gene family in Rosa chinensis and its response to salt and drought stresses
- PMID: 39161880
- PMCID: PMC11330952
- DOI: 10.1007/s13205-024-04052-0
Genome-wide identification of HSP90 gene family in Rosa chinensis and its response to salt and drought stresses
Abstract
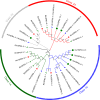
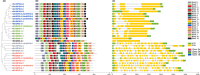
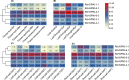
Heat shock protein 90 (HSP90) is important for many organisms, including plants. Based on the whole genome information, the gene number, gene structure, evolutionary relationship, protein structure, and active site of the HSP90 gene family in Rosa chinensis and Rubus idaeus were determined, and the expression of the HSP90 gene under salt, and drought stresses in two rose varieties Wangxifeng and Sweet Avalanche were analyzed. Six and eight HSP90 genes were identified from R. chinensis and Ru. idaeus, respectively. Phylogenetic analysis revealed that the analyzed genes were divided into two Groups and four subgroups (Classes 1a, 1b, 2a, and 2b). Although members within the same classes displayed highly similar gene structures, while the gene structures and conserved domains of Group 1 (Class 1a and 1b) and the Group 2 (Class 2a and 2b) are different. Tandem and segmental duplication genes were found in Ru. idaeus, but not in R. chinensis, perhaps explaining the difference in HSP90 gene quantity in the two analyzed species. Analysis of cis-acting elements revealed abundant abiotic stress, photolight-response, and hormone-response elements in R. chinensis HSP90s. qRT-PCR analysis suggested that RcHSP90-1-1, RcHSP90-5-1 and RcHSP90-6-1 in Sweet Avalanche and Wangxifeng varieties played important regulatory roles under salt and drought stress. The analysis of protein structure and active sites indicate that the potential different roles of RcHSP90-1-1, RcHSP90-5-1, and RcHSP90-6-1 in salt and drought stresses may come from the differences of corresponding protein structures and activation sites. These data will provide information for the breeding of rose varieties with high stress resistance.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-024-04052-0.
Keywords: Abiotic stress; HSP90 gene family; Protein active site; Relative expression; Rosa chinensis.
© The Author(s) 2024.
Figures





References
-
- Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, Kawasaki T, Shimamoto K (2010) The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7:185–196. 10.1016/j.chom.2010.02.008 10.1016/j.chom.2010.02.008 - DOI - PubMed
LinkOut - more resources
Full Text Sources

