Tracing Homopolymers in Oikopleura dioica's Mitogenome
- PMID: 39162185
- PMCID: PMC11384890
- DOI: 10.1093/gbe/evae182
Tracing Homopolymers in Oikopleura dioica's Mitogenome
Abstract
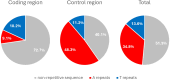
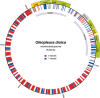
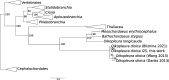
Oikopleura dioica is a planktonic tunicate (Appendicularia class) found extensively across the marine waters of the globe. The genome of a single male individual collected from Okinawa, Japan was sequenced using the single-molecule PacBio Hi-Fi method and assembled with NOVOLoci. The mitogenome is 39,268 bp long, featuring a large control region of around 22,000 bp. We annotated the proteins atp6, cob, cox1, cox2, cox3, nad1, nad4, and nad5, and found one more open reading frame that did not match any known gene. This study marks the first complete mitogenome assembly for an appendicularian, and reveals that A and T homopolymers cumulatively account for nearly half of its length. This reference sequence will be an asset for environmental DNA and phylogenetic studies.
Keywords: Appendicularia; Oikopleura dioica; homopolymers; larvacean; tunicate.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
References
-
- Bliznina A, Masunaga A, Mansfield MJ, Tan Y, Liu AW, West C, Rustagi T, Chien HC, Kumar S, Pichon J, et al. Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using nanopore-based sequencing. BMC Genomics. 2021:22(1):222. 10.1186/s12864-021-07512-6. - DOI - PMC - PubMed
-
- Danks G, Campsteijn C, Parida M, Butcher S, Doddapaneni H, Fu B, Petrin R, Metpally R, Lenhard B, Wincker P, et al. Oikobase: a genomics and developmental transcriptomics resource for the urochordate Oikopleura dioica. Nucleic Acids Res. 2013:41(D1):D845–D853. 10.1093/nar/gks1159. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

