RNA helicase D1PAS1 resolves R-loops and forms a complex for mouse pachytene piRNA biogenesis required for male fertility
- PMID: 39162228
- PMCID: PMC11514495
- DOI: 10.1093/nar/gkae712
RNA helicase D1PAS1 resolves R-loops and forms a complex for mouse pachytene piRNA biogenesis required for male fertility
Abstract
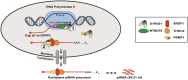
During meiosis, RNA polymerase II transcribes pachytene piRNA precursors with unusually long and unspliced transcripts from discrete autosomal loci in the mouse genome. Despite the importance of piRNA for male fertility and a well-defined maturation process, the transcriptional machinery remains poorly understood. Here, we document that D1PAS1, an ATP-dependent RNA helicase, is critical for pachytene piRNA expression from multiple genomic loci and subsequent translocation into the cytoplasm to ensure mature piRNA biogenesis. Depletion of D1PAS1 in gene-edited mice results in the accumulation of R-loops in pachytene spermatocytes, leading to DNA-damage-induced apoptosis, disruption of piRNA biogenesis, spermatogenic arrest, and male infertility. Transcriptome, genome-wide R-loop profiling, and proteomic analyses document that D1PAS1 regulates pachytene piRNA transcript elongation and termination. D1PAS1 subsequently forms a complex with nuclear export components to ensure pachytene piRNA precursor translocation from the nucleus to the cytoplasm for processing into small non-coding RNAs. Thus, our study defines D1PAS1 as a specific transcription activator that promotes R-loop unwinding and is a critical factor in pachytene piRNA biogenesis.
Published by Oxford University Press on behalf of Nucleic Acids Research 2024.
Figures







References
-
- Yanagimachi R. Knobil E., Neil J.. The Physiology of Reproduction. 1994; 2nd edn.NY: Raven Press; 189–317.
-
- Leblond C.P., Clermont Y.. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. Am. J. Anat. 1952; 90:167–215. - PubMed
-
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J.. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007; 316:744–747. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

