Evolutionary Insights from the Mitochondrial Genome of Oikopleura dioica: Sequencing Challenges, RNA Editing, Gene Transfers to the Nucleus, and tRNA Loss
- PMID: 39162337
- PMCID: PMC11384887
- DOI: 10.1093/gbe/evae181
Evolutionary Insights from the Mitochondrial Genome of Oikopleura dioica: Sequencing Challenges, RNA Editing, Gene Transfers to the Nucleus, and tRNA Loss
Abstract
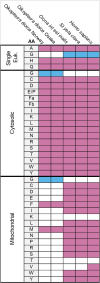
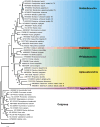
Sequencing the mitochondrial genome of the tunicate Oikopleura dioica is a challenging task due to the presence of long poly-A/T homopolymer stretches, which impair sequencing and assembly. Here, we report on the sequencing and annotation of the majority of the mitochondrial genome of O. dioica by means of combining several DNA and amplicon reads obtained by Illumina and MinIon Oxford Nanopore Technologies with public RNA sequences. We document extensive RNA editing, since all homopolymer stretches present in the mitochondrial DNA correspond to 6U-regions in the mitochondrial RNA. Out of the 13 canonical protein-coding genes, we were able to detect eight, plus an unassigned open reading frame that lacked sequence similarity to canonical mitochondrial protein-coding genes. We show that the nad3 gene has been transferred to the nucleus and acquired a mitochondria-targeting signal. In addition to two very short rRNAs, we could only identify a single tRNA (tRNA-Met), suggesting multiple losses of tRNA genes, supported by a corresponding loss of mitochondrial aminoacyl-tRNA synthetases in the nuclear genome. Based on the eight canonical protein-coding genes identified, we reconstructed maximum likelihood and Bayesian phylogenetic trees and inferred an extreme evolutionary rate of this mitochondrial genome. The phylogenetic position of appendicularians among tunicates, however, could not be accurately determined.
Keywords: Appendicularia; Oxford Nanopore Technologies; Tunicata; mtDNA; phylogenomics; poly-A/T homopolymers.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Ari Ş, Arikan M. Next-generation sequencing: advantages, disadvantages, and future. In: Hakeem KR, Tombuloğlu H, Tombuloğlu G, editors. Plant omics: trends and applications. Cham: Springer International Publishing; 2016. p. 109–135.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

