Genomic and pathogenicity analysis of two novel highly pathogenic recombinant NADC30-like PRRSV strains in China, in 2023
- PMID: 39162500
- PMCID: PMC11448138
- DOI: 10.1128/spectrum.00368-24
Genomic and pathogenicity analysis of two novel highly pathogenic recombinant NADC30-like PRRSV strains in China, in 2023
Abstract
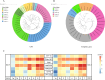
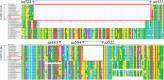
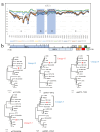
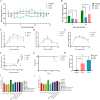
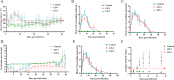
Porcine reproductive and respiratory syndrome viruses (PRRSVs) exhibit high mutability and recombination, posing challenges to their immunization and control. This study isolated two new PRRSV strains, GD-7 and GX-3, from samples collected in Guangdong and Guangxi in 2023. Whole-genome sequencing, along with phylogenetic and recombination analyses, confirmed that GD-7 and GX-3 are natural novel recombinant strains of NADC30 PRRSV. Moreover, we established a pathogenicity model for piglets and sows based on the two isolates. The results of piglet pathogenicity revealed that both GD-7 and GX-3 caused clinical symptoms such as fever, loss of appetite, depression, and slow weight gain. Moreover, we observed that the mortality rate of GD-7-inoculated group piglets was 33.3%, which was similar to that of piglets infected with other highly pathogenic PRRSV strains and exceeded the mortality rate of most NADC30-like PRRSV. In pregnant sow models, the survival rate of sows in the GD-7 group was 75%, in contrast to the GX-3 group, where no sow mortality was observed, and both strains resulted in abortion, mummified fetuses, and stillbirths. These results highlight the elevated pathogenicity of these recombinant strains in sows, with GD-7 mainly causing sows to abort, and GX-3 mainly causing sows to give birth to mummified fetuses. This study introduces two distinct clinical recombinant PRRSV strains that differ from the prevalent strains in China. This research furthers our understanding of the epidemiology of PRRSV and underscores the significance of ongoing monitoring and research in the face of evolving virus strains. Moreover, these discoveries act as early warnings, underscoring the necessity for active control and immunization against PRRSV.IMPORTANCESince the discovery of NADC30-like PRRSV in China in 2013, it has gradually become the dominant strain of PRRSV in China. NADC30-like PRRSV exhibits high recombination characteristics, constantly recombining with different strains, leading to the emergence of numerous novel strains. Of particular importance is the observation that NADC30-like PRRSV with different recombination patterns exhibits varying pathogenicity, which has a significant impact on the pig farming industry. This emphasizes the necessity of monitoring and responding to evolving PRRSV strains to develop effective immunization and control strategies. In this paper, we conducted pathogenicity studies on the isolated NADC30-like PRRSV and analyzed the differences in the genomes and pathogenicity of the different strains by recording clinical symptoms, temperature changes, detoxification tests, and changes in viremia and histopathology in infected pigs. This was done to provide a theoretical basis for the epidemiological situation and epidemic prevention and control of PRRSV.
Keywords: PRRSV; pathogenicity; phylogenetic analysis; recombination analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Pol JM, van Dijk JE, Wensvoort G, Terpstra C. 1991. Pathological, ultrastructural, and immunohistochemical changes caused by Lelystad virus in experimentally induced infections of mystery swine disease (synonym: porcine epidemic abortion and respiratory syndrome (PEARS)). Vet Q 13:137–143. doi: 10.1080/01652176.1991.9694298 - DOI - PubMed
-
- Khatun A, Park SY, Shabir N, Nazki S, Kang AR, Jeong CG, Seo BJ, Yang MS, Kim B, Seo YH, Kim WI. 2019. Evaluation of the inhibitory effects of (E)-1-(2-hydroxy-4,6-dimethoxyphenyl)-3-(naphthalen-1-yl)prop-2-en-1-one (DiNap), a natural product analog, on the replication of type 2 PRRSV in vitro and in vivo. Molecules 24:887. doi: 10.3390/molecules24050887 - DOI - PMC - PubMed
-
- Guo BQ, Chen ZS, Liu XW, Cui YZ. 1996. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus. Chin J Prev Vet Med 25:1–5.
MeSH terms
LinkOut - more resources
Full Text Sources

