Nosocomial transmission of fluconazole-resistant Candida glabrata bloodstream isolates revealed by whole-genome sequencing
- PMID: 39162519
- PMCID: PMC11448407
- DOI: 10.1128/spectrum.00883-24
Nosocomial transmission of fluconazole-resistant Candida glabrata bloodstream isolates revealed by whole-genome sequencing
Abstract
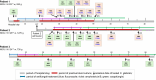
The clonal transmission of fluconazole-resistant Candida glabrata isolates within hospitals has seldom been analyzed by whole-genome sequencing (WGS). We performed WGS on 79 C. glabrata isolates, comprising 31 isolates from three premature infants with persistent C. glabrata bloodstream infection despite antifungal treatment in the same neonatal intensive care unit (NICU) in 2022 and 48 (27 fluconazole-resistant and 21 fluconazole-susceptible dose-dependent) bloodstream isolates from 48 patients in 15 South Korean hospitals from 2010 to 2022. Phylogenetic analysis based on WGS single-nucleotide polymorphisms (SNPs) distinguished the 79 isolates according to multilocus sequence typing (MLST) (17 sequence type [ST]3, 13 ST7, two ST22, 41 ST26, four ST55, and two ST59 isolates) and unveiled two possible clusters of nosocomial transmission among ST26 isolates. One cluster from two premature infants with overlapping NICU hospitalizations in 2022 encompassed 15 fluconazole-resistant isolates harboring pleiotropic drug-resistance transcription factor (Pdr1p) P258L (13 isolates) or N1086I (two isolates), together with 10 fluconazole-susceptible dose-dependent isolates lacking Pdr1p SNPs. The other cluster indicated unforeseen clonal transmission of fluconazole-resistant bloodstream isolates among five patients (four post-lung transplantation and one with diffuse interstitial lung disease) in the same hospital over 8 months. Among these five isolates, four obtained after exposure to azole antifungals harbored distinct Pdr1p SNPs (N1091D, E388Q, K365E, and R376Q). The findings reveal the transmission patterns of clonal bloodstream isolates of C. glabrata among patients undergoing antifungal treatment, exhibiting different levels of fluconazole susceptibility or distinct Pdr1p SNP profiles.
Importance: The prevalence of fluconazole-resistant bloodstream infections caused by Candida glabrata is increasing globally, but the transmission of these resistant strains within hospitals has rarely been documented. Through whole-genome sequencing and epidemiological analyses, this study identified two potential clusters of C. glabrata bloodstream infections within the same hospital, revealing the transmission of clonal C. glabrata strains with different levels of fluconazole susceptibility or distinct transcription factor pleiotropic drug resistance protein 1 (Pdr1p) single-nucleotide polymorphism profiles among patients receiving antifungal therapy.
Keywords: Candida glabrata; PDR1; clonal transmission; fluconazole resistance; multilocus sequence typing; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kwon YJ, Won EJ, Jeong SH, Shin KS, Shin JH, Kim YR, Kim HS, Kim YA, Uh Y, Kim TS, Park JH, Lee J, Choi MJ, Byun SA, Kim SH, Shin JH. 2021. Dynamics and predictors of mortality due to candidemia caused by different Candida species: comparison of intensive care unit-associated candidemia (ICUAC) and non-ICUAC. J Fungi (Basel) 7:597. doi:10.3390/jof7080597 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

