Bacterial Genomics for National Antimicrobial Resistance Surveillance in Cambodia
- PMID: 39163245
- PMCID: PMC12526891
- DOI: 10.1093/infdis/jiae417
Bacterial Genomics for National Antimicrobial Resistance Surveillance in Cambodia
Abstract
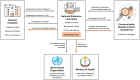
Background: Antimicrobial resistance (AMR) surveillance in low- and middle-income countries (LMICs) often relies on poorly resourced laboratory processes. Centralized sequencing was combined with cloud-based, open-source bioinformatics solutions for national AMR surveillance in Cambodia.
Methods: Blood cultures growing gram-negative bacteria were collected at 6 Cambodian hospitals (January 2021 to October 2022). Isolates were obtained from pure plate growth and shotgun DNA sequencing performed in country. Using public nucleotide and protein databases, reads were aligned for pathogen identification and AMR gene characterization. Multilocus sequence typing was performed on whole-genome assemblies and haplotype clusters compared against published genomes.
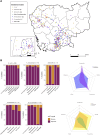
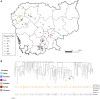
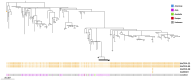
Results: Genes associated with acquired resistance to fluoroquinolones were identified in 59%, trimethoprim/sulfamethoxazole in 45%, and aminoglycosides in 52% of 715 isolates. Extended-spectrum β-lactamase encoding genes were identified in 34% isolates, most commonly blaCTX-M-15, blaCTX-M-27, and blaCTX-M-55 in Escherichia coli sequence types 131 and 1193. Carbapenemase genes were identified in 12% isolates, most commonly blaOXA-23, blaNDM-1, blaOXA-58, and blaOXA-66 in Acinetobacter species. Phylogenetic analysis revealed clonal strains of Acinetobacter baumannii, representing suspected nosocomial outbreaks, and genetic clusters of quinolone-resistant typhoidal Salmonella and extended-spectrum β-lactamase E. coli cases suggesting community transmission.
Conclusions: With accessible sequencing platforms and bioinformatics solutions, bacterial genomics can supplement AMR surveillance in LMICs.
Keywords: Cambodia; bacterial resistance; genomic surveillance; outbreak investigation; resistance genes.
Published by Oxford University Press on behalf of Infectious Diseases Society of America 2024.
Conflict of interest statement
Potential conflicts of interest . Since this work was completed, J. M. has accepted full-time employment with Sanofi Pasteur. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 2013; 13:1057–98. - PubMed
-
- World Health Organization . Global action plan on antimicrobial resistance. 2015. https://www.who.int/publications/i/item/9789241509763. Accessed 22 March 2023. - PubMed
-
- World Health Organization . Global antimicrobial resistance and use surveillance system (GLASS) report: 2022. 2023. https://www.who.int/publications/i/item/9789240062702. Accessed 22 March 2023.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

