An engineered NKp46 antibody for construction of multi-specific NK cell engagers
- PMID: 39163262
- PMCID: PMC11359164
- DOI: 10.1093/protein/gzae013
An engineered NKp46 antibody for construction of multi-specific NK cell engagers
Abstract
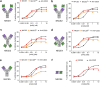
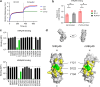
Recent developments in cancer immunotherapy have highlighted the potential of harnessing natural killer (NK) cells in the treatment of neoplastic malignancies. Of these, bispecific antibodies, and NK cell engager (NKCE) protein therapeutics in particular, have been of interest. Here, we used phage display and yeast surface display to engineer RLN131, a unique cross-reactive antibody that binds to human, mouse, and cynomolgus NKp46, an activating receptor found on NK cells. RLN131 induced proliferation and activation of primary NK cells, and was used to create bispecific NKCE constructs of varying configurations and valency. All NKCEs were able to promote greater NK cell cytotoxicity against tumor cells than an unmodified anti-CD20 monoclonal antibody, and activity was observed irrespective of whether the constructs contained a functional Fc domain. Competition binding and fine epitope mapping studies were used to demonstrate that RLN131 binds to a conserved epitope on NKp46, underlying its species cross-reactivity.
Keywords: NCR1; NK cell engager; NKp46; antibody engineering; bispecific antibody; species cross reactive.
© The Author(s) 2024. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
R.B.L. and J.R.C. are inventors on intellectual property related to this work that is owned by Stanford University. C.L.M. holds equity in and consults for CARGO Therapeutics, Link Cell Therapies and Ensoma, received research funding from Lyell Immunopharma and Tune therapeutics, and consults for Immatics. J.B.S. holds equity in and consults for Indapta Therapeutics. J.R.C. is a cofounder and equity holder of Combangio, Inc. (now Kala Bio), xCella Biosciences (now OmniAb), Charged Biotherapeutics, TwoStep Therapeutics, and Red Tree Venture Capital, has financial interests in Aravive, Inc., is a member of the Board of Directors of OmniAb, Revel Pharmaceuticals, Excellergy Therapeutics, Rondo Therapeutics, Tachyon Therapeutics, and Biograph 55, and is a Board Observer at Acrigen Biosciences. The other authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

