Rapidly evolving genome and epigenome editing technologies
- PMID: 39163859
- PMCID: PMC11403209
- DOI: 10.1016/j.ymthe.2024.08.011
Rapidly evolving genome and epigenome editing technologies
Abstract
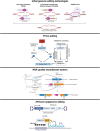
Genome editing technologies are rapidly evolving, from the early zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 (Figure 1, initial genome editing technologies), which generate double-strand breaks (DSBs), to base editing, which makes precise nucleobase conversion without inducing DSBs, and prime editing, which can carry out all types of edits without DSBs or donor DNA templates. The emergence of these revolutionary technologies offers us unprecedented opportunities for biomedical research and therapy development.
Copyright © 2024 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Levesque S., Cosentino A., Verma A., Genovese P., Bauer D.E. Enhancing prime editing in hematopoietic stem and progenitor cells by modulating nucleotide metabolism. Nat. Biotechnol. 2024;41:1446–1456. - PubMed
-
- Yu G., Kim H.K., Park J., Kwak H., Cheong Y., Kim D., Kim J., Kim J., Kim H.H. Prediction of efficiencies for diverse prime editing systems in multiple cell types. Cell. 2023;186:2256–2272.e23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

