Fish-hunting cone snail disrupts prey's glucose homeostasis with weaponized mimetics of somatostatin and insulin
- PMID: 39164229
- PMCID: PMC11336141
- DOI: 10.1038/s41467-024-50470-2
Fish-hunting cone snail disrupts prey's glucose homeostasis with weaponized mimetics of somatostatin and insulin
Abstract
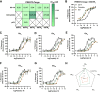
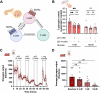
Venomous animals have evolved diverse molecular mechanisms to incapacitate prey and defend against predators. Most venom components disrupt nervous, locomotor, and cardiovascular systems or cause tissue damage. The discovery that certain fish-hunting cone snails use weaponized insulins to induce hypoglycemic shock in prey highlights a unique example of toxins targeting glucose homeostasis. Here, we show that, in addition to insulins, the deadly fish hunter, Conus geographus, uses a selective somatostatin receptor 2 (SSTR2) agonist that blocks the release of the insulin-counteracting hormone glucagon, thereby exacerbating insulin-induced hypoglycemia in prey. The native toxin, Consomatin nG1, exists in several proteoforms with a minimized vertebrate somatostatin-like core motif connected to a heavily glycosylated N-terminal region. We demonstrate that the toxin's N-terminal tail closely mimics a glycosylated somatostatin from fish pancreas and is crucial for activating the fish SSTR2. Collectively, these findings provide a stunning example of chemical mimicry, highlight the combinatorial nature of venom components, and establish glucose homeostasis as an effective target for prey capture.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

