Mdm2 requires Sprouty4 to regulate focal adhesion formation and metastasis independent of p53
- PMID: 39164253
- PMCID: PMC11336179
- DOI: 10.1038/s41467-024-51488-2
Mdm2 requires Sprouty4 to regulate focal adhesion formation and metastasis independent of p53
Abstract
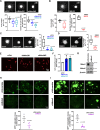
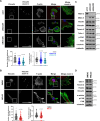
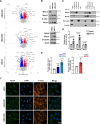
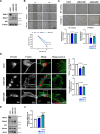
Although the E3 ligase Mdm2 and its homologue and binding partner MdmX are the major regulators of the p53 tumor suppressor protein, it is now evident that Mdm2 and MdmX have multiple functions that do not involve p53. As one example, it is known that Mdm2 can regulate cell migration, although mechanistic insight into this function is still lacking. Here we show in cells lacking p53 expression that knockdown of Mdm2 or MdmX, as well as pharmacological inhibition of the Mdm2/MdmX complex, not only reduces cell migration and invasion, but also impairs cell spreading and focal adhesion formation. In addition, Mdm2 knockdown decreases metastasis in vivo. Interestingly, Mdm2 downregulates the expression of Sprouty4, which is required for the Mdm2 mediated effects on cell migration, focal adhesion formation and metastasis. Further, our findings indicate that Mdm2 dampening of Sprouty4 is a prerequisite for maintaining RhoA levels in the cancer cells that we have studied. Taken together we describe a molecular mechanism whereby the Mdm2/MdmX complex through Sprouty4 regulates cellular processes leading to increase metastatic capability independently of p53.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Iwakuma, T. & Lozano, G. MDM2, an introduction. Mol. Cancer Res.1, 993–1000 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

