FOXO-regulated OSER1 reduces oxidative stress and extends lifespan in multiple species
- PMID: 39164296
- PMCID: PMC11336091
- DOI: 10.1038/s41467-024-51542-z
FOXO-regulated OSER1 reduces oxidative stress and extends lifespan in multiple species
Abstract
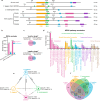
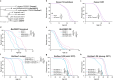
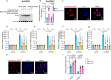
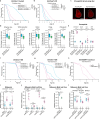
FOXO transcription factors modulate aging-related pathways and influence longevity in multiple species, but the transcriptional targets that mediate these effects remain largely unknown. Here, we identify an evolutionarily conserved FOXO target gene, Oxidative stress-responsive serine-rich protein 1 (OSER1), whose overexpression extends lifespan in silkworms, nematodes, and flies, while its depletion correspondingly shortens lifespan. In flies, overexpression of OSER1 increases resistance to oxidative stress, starvation, and heat shock, while OSER1-depleted flies are more vulnerable to these stressors. In silkworms, hydrogen peroxide both induces and is scavenged by OSER1 in vitro and in vivo. Knockdown of OSER1 in Caenorhabditis elegans leads to increased ROS production and shorter lifespan, mitochondrial fragmentation, decreased ATP production, and altered transcription of mitochondrial genes. Human proteomic analysis suggests that OSER1 plays roles in oxidative stress response, cellular senescence, and reproduction, which is consistent with the data and suggests that OSER1 could play a role in fertility in silkworms and nematodes. Human studies demonstrate that polymorphic variants in OSER1 are associated with human longevity. In summary, OSER1 is an evolutionarily conserved FOXO-regulated protein that improves resistance to oxidative stress, maintains mitochondrial functional integrity, and increases lifespan in multiple species. Additional studies will clarify the role of OSER1 as a critical effector of healthy aging.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

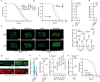


References
-
- van der Horst, A. & Burgering, B. M. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol.8, 440–450 (2007). - PubMed
-
- Kenyon, C. J. The genetics of ageing. Nature464, 504–512 (2010). - PubMed
-
- Greer, E. L. et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem.282, 30107–30119 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

