Modeling relaxation experiments with a mechanistic model of gene expression
- PMID: 39164646
- PMCID: PMC11334594
- DOI: 10.1186/s12859-024-05816-4
Modeling relaxation experiments with a mechanistic model of gene expression
Abstract
Background: In the present work, we aimed at modeling a relaxation experiment which consists in selecting a subfraction of a cell population and observing the speed at which the entire initial distribution for a given marker is reconstituted.
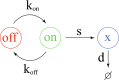
Methods: For this we first proposed a modification of a previously published mechanistic two-state model of gene expression to which we added a state-dependent proliferation term. This results in a system of two partial differential equations. Under the assumption of a linear dependence of the proliferation rate with respect to the marker level, we could derive the asymptotic profile of the solutions of this model.
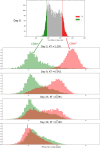
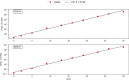
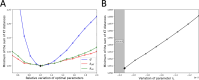
Results: In order to confront our model with experimental data, we generated a relaxation experiment of the CD34 antigen on the surface of TF1-BA cells, starting either from the highest or the lowest CD34 expression levels. We observed in both cases that after approximately 25 days the distribution of CD34 returns to its initial stationary state. Numerical simulations, based on parameter values estimated from the dataset, have shown that the model solutions closely align with the experimental data from the relaxation experiments.
Conclusion: Altogether our results strongly support the notion that cells should be seen and modeled as probabilistic dynamical systems.
Keywords: Asymptotic profile; Relaxation experiments; Two-state model.
© 2024. The Author(s).
Conflict of interest statement
None
Figures


References
-
- Kupiec JJ. A probabilistic theory for cell differentiation, embryonic mortality and DNA c-value paradox. Specul Sci Technol. 1983;6(5):471–8.
-
- Schrödinger E. What is life? The physical aspect of the living cell. Cambridge: Cambridge University Press; 1944.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

