Epigenetic patterns, accelerated biological aging, and enhanced epigenetic drift detected 6 months following COVID-19 infection: insights from a genome-wide DNA methylation study
- PMID: 39164752
- PMCID: PMC11337605
- DOI: 10.1186/s13148-024-01724-9
Epigenetic patterns, accelerated biological aging, and enhanced epigenetic drift detected 6 months following COVID-19 infection: insights from a genome-wide DNA methylation study
Erratum in
-
Correction: Epigenetic patterns, accelerated biological aging, and enhanced epigenetic drift detected 6 months following COVID‑19 infection: insights from a genome‑wide DNA methylation study.Clin Epigenetics. 2024 Nov 14;16(1):158. doi: 10.1186/s13148-024-01764-1. Clin Epigenetics. 2024. PMID: 39543717 Free PMC article. No abstract available.
Abstract
Background: The epigenetic status of patients 6-month post-COVID-19 infection remains largely unexplored. The existence of long-COVID, or post-acute sequelae of SARS-CoV-2 infection (PASC), suggests potential long-term changes. Long-COVID includes symptoms like fatigue, neurological issues, and organ-related problems, regardless of initial infection severity. The mechanisms behind long-COVID are unclear, but virus-induced epigenetic changes could play a role.
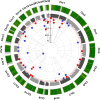
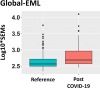
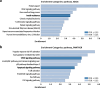
Methods and results: Our study explores the lasting epigenetic impacts of SARS-CoV-2 infection. We analyzed genome-wide DNA methylation patterns in an Italian cohort of 96 patients 6 months after COVID-19 exposure, comparing them to 191 healthy controls. We identified 42 CpG sites with significant methylation differences (FDR < 0.05), primarily within CpG islands and gene promoters. Dysregulated genes highlighted potential links to glutamate/glutamine metabolism, which may be relevant to PASC symptoms. Key genes with potential significance to COVID-19 infection and long-term effects include GLUD1, ATP1A3, and ARRB2. Furthermore, Horvath's epigenetic clock showed a slight but significant age acceleration in post-COVID-19 patients. We also observed a substantial increase in stochastic epigenetic mutations (SEMs) in the post-COVID-19 group, implying potential epigenetic drift. SEM analysis identified 790 affected genes, indicating dysregulation in pathways related to insulin resistance, VEGF signaling, apoptosis, hypoxia response, T-cell activation, and endothelin signaling.
Conclusions: Our study provides valuable insights into the epigenetic consequences of COVID-19. Results suggest possible associations with accelerated aging, epigenetic drift, and the disruption of critical biological pathways linked to insulin resistance, immune response, and vascular health. Understanding these epigenetic changes could be crucial for elucidating the complex mechanisms behind long-COVID and developing targeted therapeutic interventions.
Keywords: COVID-19; DNA methylation; EWAS; Epigenetic clock; Epigenetic drift; PASC; SARS-CoV-2; Stochastic epigenetic mutation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

