Targeting circ-0034880-enriched tumor extracellular vesicles to impede SPP1highCD206+ pro-tumor macrophages mediated pre-metastatic niche formation in colorectal cancer liver metastasis
- PMID: 39164758
- PMCID: PMC11334400
- DOI: 10.1186/s12943-024-02086-9
Targeting circ-0034880-enriched tumor extracellular vesicles to impede SPP1highCD206+ pro-tumor macrophages mediated pre-metastatic niche formation in colorectal cancer liver metastasis
Abstract
Background: Information transmission between primary tumor cells and immunocytes or stromal cells in distal organs is a critical factor in the formation of pre-metastatic niche (PMN). Understanding this mechanism is essential for developing effective therapeutic strategy against tumor metastasis. Our study aims to prove the hypothesis that circ-0034880-enriched tumor-derived extracellular vesicles (TEVs) mediate the formation of PMN and colorectal cancer liver metastasis (CRLM), and targeting circ-0034880-enriched TEVs might be an effective therapeutic strategy against PMN formation and CRLM.
Methods: We utilized qPCR and FISH to measure circRNAs expression levels in human CRC plasma, primary CRC tissues, and liver metastatic tissues. Additionally, we employed immunofluorescence, RNA sequencing, and in vivo experiments to assess the effect mechanism of circ-0034880-enriched TEVs on PMN formation and CRC metastasis. DARTS, CETSA and computational docking modeling were applied to explore the pharmacological effects of Ginsenoside Rb1 in impeding PMN formation.
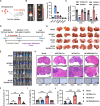
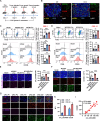
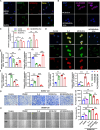
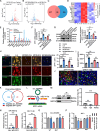
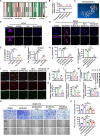
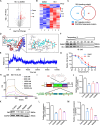
Results: We found that circ-0034880 was highly enriched in plasma extracellular vesicles (EVs) derived from CRC patients and closely associated with CRLM. Functionally, circ-0034880-enriched TEVs entered the liver tissues and were absorbed by macrophages in the liver through bloodstream. Mechanically, TEVs-released circ-0034880 enhanced the activation of SPP1highCD206+ pro-tumor macrophages, reshaping the metastasis-supportive host stromal microenvironment and promoting overt metastasis. Importantly, our mechanistic findings led us to discover that the natural product Ginsenoside Rb1 impeded the activation of SPP1highCD206+ pro-tumor macrophages by reducing circ-0034880 biogenesis, thereby suppressing PMN formation and inhibiting CRLM.
Conclusions: Circ-0034880-enriched TEVs facilitate strong interaction between primary tumor cells and SPP1highCD206+ pro-tumor macrophages, promoting PMN formation and CRLM. These findings suggest the potential of using Ginsenoside Rb1 as an alternative therapeutic agent to reshape PMN formation and prevent CRLM.
Keywords: Circ-0034880; Extracellular vesicles; Ginsenoside Rb1; Pro-tumor macrophages; Tumor metastasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Wille-Jørgensen P, Syk I, Smedh K, et al. Effect of More vs Less Frequent Follow-up Testing on Overall and Colorectal Cancer-Specific Mortality in Patients With Stage II or III Colorectal Cancer: The COLOFOL Randomized Clinical Trial. JAMA. 2018;319:2095–103. 10.1001/jama.2018.5623. 10.1001/jama.2018.5623 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

