The DNA repair pathway as a therapeutic target to synergize with trastuzumab deruxtecan in HER2-targeted antibody-drug conjugate-resistant HER2-overexpressing breast cancer
- PMID: 39164784
- PMCID: PMC11337831
- DOI: 10.1186/s13046-024-03143-3
The DNA repair pathway as a therapeutic target to synergize with trastuzumab deruxtecan in HER2-targeted antibody-drug conjugate-resistant HER2-overexpressing breast cancer
Abstract
Background: Anti-HER2 therapies, including the HER2 antibody-drug conjugates (ADCs) trastuzumab emtansine (T-DM1) and trastuzumab deruxtecan (T-DXd), have led to improved survival outcomes in patients with HER2-overexpressing (HER2+) metastatic breast cancer. However, intrinsic or acquired resistance to anti-HER2-based therapies remains a clinical challenge in these patients, as there is no standard of care following disease progression. The purpose of this study was to elucidate the mechanisms of resistance to T-DM1 and T-DXd in HER2+ BC patients and preclinical models and identify targets whose inhibition enhances the antitumor activity of T-DXd in HER2-directed ADC-resistant HER2+ breast cancer in vitro and in vivo.
Methods: Targeted DNA and whole transcriptome sequencing were performed in breast cancer patient tissue samples to investigate genetic aberrations that arose after anti-HER2 therapy. We generated T-DM1 and T-DXd-resistant HER2+ breast cancer cell lines. To elucidate their resistance mechanisms and to identify potential synergistic kinase targets for enhancing the efficacy of T-DXd, we used fluorescence in situ hybridization, droplet digital PCR, Western blotting, whole-genome sequencing, cDNA microarray, and synthetic lethal kinome RNA interference screening. In addition, cell viability, colony formation, and xenograft assays were used to determine the synergistic antitumor effect of T-DXd combinations.
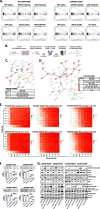
Results: We found reduced HER2 expression in patients and amplified DNA repair-related genes in patients after anti-HER2 therapy. Reduced ERBB2 gene amplification in HER2-directed ADC-resistant HER2+ breast cancer cell lines was through DNA damage and epigenetic mechanisms. In HER2-directed ADC-resistant HER2+ breast cancer cell lines, our non-biased RNA interference screening identified the DNA repair pathway as a potential target within the canonical pathways to enhance the efficacy of T-DXd. We validated that the combination of T-DXd with ataxia telangiectasia and Rad3-related inhibitor, elimusertib, led to significant breast cancer cell death in vitro (P < 0.01) and in vivo (P < 0.01) compared to single agents.
Conclusions: The DNA repair pathways contribute to HER2-directed ADC resistance. Our data justify exploring the combination treatment of T-DXd with DNA repair-targeting drugs to treat HER2-directed ADC-resistant HER2+ breast cancer in clinical trials.
Keywords: DNA damage repair pathway; HER2 antibody–drug conjugates; HER2+ breast cancer; HER2-directed ADC resistance; T-DXd.
© 2024. The Author(s).
Conflict of interest statement
Naoto T. Ueno and Jangsoon Lee have contracted research with Daiichi Sankyo. Seock-Ah Im reports advisory role for AstraZeneca, Daiichi-Sankyo, Eisai, Hanmi, Idience, Lilly, MSD, Novartis, Pfizer, Bertis, and Roche and has received research grants through her institution from AstraZeneca, Boryung Pharm, Daiichi-Sankyo, Daewoong Pharm, Eisai, Pfizer, and Roche. All other authors declare no competing interests.
Figures




References
-
- Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353(16):1652–4. - PubMed
-
- Payne SJ, Bowen RL, Jones JL, Wells CA. Predictive markers in breast cancer–the present. Histopathology. 2008;52(1):82–90. - PubMed
-
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9(1):16–32. - PubMed
-
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–92. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

