WNT5A is a putative epi-driver of prostate cancer metastasis to the bone
- PMID: 39164966
- PMCID: PMC11335815
- DOI: 10.1002/cam4.70122
WNT5A is a putative epi-driver of prostate cancer metastasis to the bone
Abstract
Background: Current diagnostic tools are unable to distinguish low-grade indolent prostate cancer (PrCa) from that with a propensity to become metastatic and/or lethal. Recent evidence suggests that reprogramming of the transcriptome may drive the metastatic phenotype, and that this reprogramming is controlled, at least in part, by epigenetic changes to the DNA of cancer cells, including methylation. These changes, referred to as 'epigenetic drivers,' have previously been associated with cancer cell survival.
Methods: Here, using Illumina Methylation EPIC array data of paired primary PrCa and metastatic bone samples, we identified WNT5A as a putative epi-driver of PrCa metastasis to the bone, which was further validated in vitro.
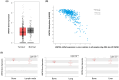
Results: Significantly higher WNT5A methylation was observed in primary PrCa samples and 22Rv1 cells compared to metastatic bone samples and PC-3 cells. This higher methylation was associated with significantly lower WNT5A gene expression.
Conclusion: Given the limited effective therapies available for metastatic cancer sufferers, particularly those whose disease has metastasised to the bone, WNT5A presents as a potential putative target for therapy.
Keywords: DNA methylation; epigenetic driver; gene expression; metastasis; prostate cancer.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Howlader N, Noone AM, Krapcho M, et al. (eds) SEER cancer statistics review, 1975‐2017. National Cancer Institute; 2020. https://seer.cancer.gov/csr/1975_2017/
-
- Gandaglia G, Abdollah F, Schiffmann J, et al. Distribution of metastatic sites in patients with prostate cancer: a population‐based analysis. Prostate. 2014;74(2):210‐216. - PubMed
-
- Nørgaard M, Jensen A, Jacobsen JB, Cetin K, Fryzek JP, Sørensen HT. Skeletal related events, bone metastasis and survival of prostate cancer: a population based cohort study in Denmark (1999 to 2007). J Urol. 2010;184(1):162‐167. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

