Broadening sarbecovirus neutralization with bispecific antibodies combining distinct conserved targets on the receptor binding domain
- PMID: 39165108
- PMCID: PMC11340772
- DOI: 10.1080/21645515.2024.2388344
Broadening sarbecovirus neutralization with bispecific antibodies combining distinct conserved targets on the receptor binding domain
Abstract
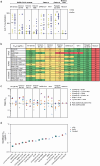
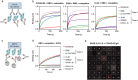
Monoclonal neutralizing antibodies (mAbs) are considered an important prophylactic against SARS-CoV-2 infection in at-risk populations and a strategy to counteract future sarbecovirus-induced disease. However, most mAbs isolated so far neutralize only a few sarbecovirus strains. Therefore, there is a growing interest in bispecific antibodies (bsAbs) which can simultaneously target different spike epitopes and thereby increase neutralizing breadth and prevent viral escape. Here, we generate and characterize a panel of 30 novel broadly reactive bsAbs using an efficient controlled Fab-arm exchange protocol. We specifically combine some of the broadest mAbs described so far, which target conserved epitopes on the receptor binding domain (RBD). Several bsAbs show superior cross-binding and neutralization compared to the parental mAbs and cocktails against sarbecoviruses from diverse clades, including recent SARS-CoV-2 variants. BsAbs which include mAb COVA2-02 are among the most potent and broad combinations. As a result, we study the unknown epitope of COVA2-02 and show that this mAb targets a distinct conserved region at the base of the RBD, which could be of interest when designing next-generation bsAb constructs to contribute to a better pandemic preparedness.
Keywords: SARS-CoV-2; bispecific antibodies; breadth; cross-reactivity; neutralization; sarbecoviruses; variants.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi:10.1126/science.abc5902. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
