Factors governing attachment of Rhizobium leguminosarum to legume roots at acid, neutral, and alkaline pHs
- PMID: 39166858
- PMCID: PMC11406972
- DOI: 10.1128/msystems.00422-24
Factors governing attachment of Rhizobium leguminosarum to legume roots at acid, neutral, and alkaline pHs
Abstract
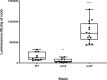
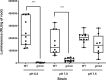
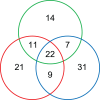
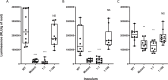
Rhizobial attachment to host legume roots is the first physical interaction of bacteria and plants in symbiotic nitrogen fixation. The pH-dependent primary attachment of Rhizobium leguminosarum biovar viciae 3841 to Pisum sativum (pea) roots was investigated by genome-wide insertion sequencing, luminescence-based attachment assays, and proteomic analysis. Under acid, neutral, or alkaline pH, a total of 115 genes are needed for primary attachment under one or more environmental pH, with 22 genes required for all. These include components of cell surfaces and membranes, together with enzymes that construct and modify them. Mechanisms of dealing with stress also play a part; however, exact requirements vary depending on environmental pH. RNASeq showed that knocking out the two transcriptional regulators required for attachment causes massive changes in the bacterial cell surface. Approximately half of the 54 proteins required for attachment at pH 7.0 have a role in the later stages of nodule formation. We found no evidence for a single rhicadhesin responsible for alkaline attachment, although sonicated cell surface fractions inhibited root attachment at alkaline pH. Our results demonstrate the complexity of primary root attachment and illustrate the diversity of mechanisms involved.
Importance: The first step by which bacteria interact with plant roots is by attachment. In this study, we use a combination of insertion sequencing and biochemical analysis to determine how bacteria attach to pea roots and how this is influenced by pH. We identify several key adhesins, which are molecules that enable bacteria to stick to roots. This includes a novel filamentous hemagglutinin which is needed at all pHs for attachment. Overall, 115 proteins are required for attachment at one or more pHs.
Keywords: adhesin; attachment; legume; pea; rhizobium; root.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

