Essential genes for Haemophilus parainfluenzae survival and biofilm growth
- PMID: 39166876
- PMCID: PMC11406952
- DOI: 10.1128/msystems.00674-24
Essential genes for Haemophilus parainfluenzae survival and biofilm growth
Abstract
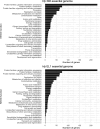
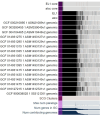
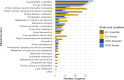
Haemophilus parainfluenzae (Hp) is a Gram-negative, highly prevalent, and abundant commensal in the human oral cavity, and an infrequent extraoral opportunistic pathogen. Hp occupies multiple niches in the oral cavity, including the supragingival plaque biofilm. Little is known about how Hp interacts with its neighbors in healthy biofilms nor its mechanisms of pathogenesis as an opportunistic pathogen. To address this, we identified the essential genome and conditionally essential genes in in vitro biofilms aerobically and anaerobically. Using transposon insertion sequencing (TnSeq) with a highly saturated mariner transposon library in two strains, the ATCC33392 type-strain (Hp 392) and oral isolate EL1 (Hp EL1), we show that the essential genomes of Hp 392 and Hp EL1 are composed of 395 (20%) and 384 (19%) genes, respectively. The core essential genome, consisting of 341 (17%) essential genes conserved between both strains, was composed of genes associated with genetic information processing, carbohydrate, protein, and energy metabolism. We also identified conditionally essential genes for aerobic and anaerobic biofilm growth, which were associated with carbohydrate and energy metabolism in both strains. RNAseq analysis determined that most genes upregulated during anaerobic growth are not essential for Hp 392 anaerobic survival. The completion of this library and analysis under these conditions gives us a foundational insight into the basic biology of H. parainfluenzae in differing oxygen conditions, similar to its in vivo habitat. This library presents a valuable tool for investigation into conditionally essential genes for an organism that lives in close contact with many microbial species in the human oral habitat.IMPORTANCEHaemophilus parainfluenzae is a highly abundant human commensal microbe, present in most healthy individuals where it colonizes the mouth. H. parainfluenzae correlates with good oral health and may play a role in preservation of healthy host status. Also, H. parainfluenzae can cause opportunistic infections outside of the oral cavity. To date, little is known about how H. parainfluenzae colonizes the human host, despite being such a frequent and abundant part of our human microbiome. Here, we demonstrate the creation and use of a powerful tool, a TnSeq library, used to identify genes necessary for both the outright growth of this organism and also genes conditionally essential for growth in varying oxygen status which it can encounter in the human host. This tool and these data serve as a foundation for further study of this relatively unknown organism that may play a role in preserving human health.
Keywords: Haemophilus; TnSeq; biofilms; commensal; dental plaque; facultative anaerobes; oral microbiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
Essential genes for Haemophilus parainfluenzae survival and biofilm growth.bioRxiv [Preprint]. 2024 Mar 31:2024.03.31.587483. doi: 10.1101/2024.03.31.587483. bioRxiv. 2024. Update in: mSystems. 2024 Sep 17;9(9):e0067424. doi: 10.1128/msystems.00674-24. PMID: 38585970 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
