Genomic landscape of gallbladder cancer: insights from whole exome sequencing
- PMID: 39166960
- PMCID: PMC11573093
- DOI: 10.1097/JS9.0000000000002031
Genomic landscape of gallbladder cancer: insights from whole exome sequencing
Abstract
Background: Gallbladder cancer (GBC) is a common gastrointestinal malignancy noted for its aggressive characteristics and poor prognosis, which is mostly caused by delayed detection. However, the scarcity of information regarding somatic mutations in Indian patients with GBC has hampered the development of efficient therapeutic options. In the present study, the authors attempted to bridge this gap by revealing the mutational profile of GBC.
Materials and methods: To evaluate the somatic mutation profile, whole exome sequencing (WES) was performed on 66 tumor and matched blood samples from individuals with GBC. Somatic variant calling was performed using GATK pipeline. Variants were annotated at pathogenic and oncogenic levels, using ANNOVAR, VEP tools and the OncoKB database. Mutational signature analysis, oncogenic pathway analysis and cancer driver genes identification were performed at the functional level by using the maftools package.
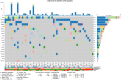
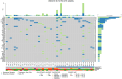
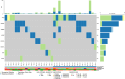
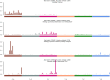
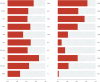
Results: Our findings focused on the eight most altered genes with pathogenic and oncogenic mutations: TP53, SMAD4, ERBB3, KRAS, ARID1A, PIK3CA, RB1, and AXIN1. Genes with pathogenic single nucleotide variations (SNVs) were enriched in oncogenic signaling pathways, particularly RTK-RAS, WNT, and TP53 pathways. Furthermore, our research related certain mutational signatures, such as cosmic 1, cosmic 6, and cosmic 18, 29, to known characteristics including patient age and tobacco smoking, providing important insights into disease etiology.
Conclusions: Given the scarcity of exome-based sequencing studies focusing on the Indian population, this study represents a significant step forward in providing a framework for additional in-depth mutational analysis. Genes with substantial oncogenic and pathogenic mutations are promising candidates for developing targeted mutation panels, particularly for GBC detection.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors declare that they have no financial conflict of interest with regard to the content of this report.
Figures


References
-
- Valle J, Wasan H, Palmer DH, et al. . Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273–1281. - PubMed
-
- Butti AK, Yadav SK, Verma A, et al. . Chronic calculus cholecystitis: is histopathology essential post-cholecystectomy? Indian J Cancer 2020;57:89–92. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

