Transcriptional network dynamics in early T cell development
- PMID: 39167073
- PMCID: PMC11338287
- DOI: 10.1084/jem.20230893
Transcriptional network dynamics in early T cell development
Abstract
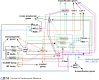
The rate at which cells enter the T cell pathway depends not only on the immigration of hematopoietic precursors into the strong Notch signaling environment of the thymus but also on the kinetics with which each individual precursor cell reaches T-lineage commitment once it arrives. Notch triggers a complex, multistep gene regulatory network in the cells in which the steps are stereotyped but the transition speeds between steps are variable. Progenitor-associated transcription factors delay T-lineage differentiation even while Notch-induced transcription factors within the same cells push differentiation forward. Progress depends on regulator cross-repression, on breaching chromatin barriers, and on shifting, competitive collaborations between stage-specific and stably expressed transcription factors, as reviewed here.
© 2024 Shin et al.
Conflict of interest statement
Disclosures: E.V. Rothenberg was on the Scientific Advisory Board for Century Therapeutics, Inc. until January 2023. The advisory work that she did for them does not have any overlap with material in the present article. The other authors have no disclosures to report.
Figures



References
-
- Alonzo, E.S., Gottschalk R.A., Das J., Egawa T., Hobbs R.M., Pandolfi P.P., Pereira P., Nichols K.E., Koretzky G.A., Jordan M.S., et al. 2010. Development of promyelocytic zinc finger and ThPOK-expressing innate γδ T cells is controlled by strength of TCR signaling and Id3. J. Immunol. 184:1268–1279. 10.4049/jimmunol.0903218 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

