Somatic mutations in tumor-infiltrating lymphocytes impact on antitumor immunity
- PMID: 39167601
- PMCID: PMC11363295
- DOI: 10.1073/pnas.2320189121
Somatic mutations in tumor-infiltrating lymphocytes impact on antitumor immunity
Abstract
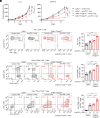
Immune checkpoint inhibitors (ICIs) exert clinical efficacy against various types of cancers by reinvigorating exhausted CD8+ T cells that can expand and directly attack cancer cells (cancer-specific T cells) among tumor-infiltrating lymphocytes (TILs). Although some reports have identified somatic mutations in TILs, their effect on antitumor immunity remains unclear. In this study, we successfully established 18 cancer-specific T cell clones, which have an exhaustion phenotype, from the TILs of four patients with melanoma. We conducted whole-genome sequencing for these T cell clones and identified various somatic mutations in them with high clonality. Among the somatic mutations, an SH2D2A loss-of-function frameshift mutation and TNFAIP3 deletion could activate T cell effector functions in vitro. Furthermore, we generated CD8+ T cell-specific Tnfaip3 knockout mice and showed that Tnfaip3 function loss in CD8+ T cell increased antitumor immunity, leading to remarkable response to PD-1 blockade in vivo. In addition, we analyzed bulk CD3+ T cells from TILs in additional 12 patients and identified an SH2D2A mutation in one patient through amplicon sequencing. These findings suggest that somatic mutations in TILs can affect antitumor immunity and suggest unique biomarkers and therapeutic targets.
Keywords: T cell; cancer immunology; somatic mutation; tumor-infiltrating lymphocytes.
Conflict of interest statement
Competing interests statement:K. Yamashita is an employee of KOTAI Biotechnologies. T. Inozume received honoraria from Ono Pharmaceutical, Bristol-Myers Squibb, and MSD outside of this study. H. Mano is a board member of CureGene; and received honoraria from Konica Minolta outside of this study. S. Toyooka received honoraria from TAIHO PHARMA, Eli Lilly, Chugai Pharmaceutical, GUARDANT, AstraZeneca, Illumina, MERCK, CSL Behring, Johnson & Johnson, Nippon Kayaku, INTUITIVE, Daiichi-Sankyo, ONO PHARMACEUTICAL, Medtronic, Ziosoft, NOVARTIS, Sysmex, and Riken Genesis outside of this study. Y. Togashi received honoraria from Ono Pharmaceutical and Bristol-Myers Squibb, AstraZeneca, Chugai Pharmaceutical, and MSD outside of this study. H. Mano received research grants from Daiichi Sankyo, Konica Minolta, Ambry Genetics, Ono Pharmaceutical and PFDeNA outside of this study. S. Toyooka received research grants from TAIHO PHARMA, Eli Lilly, Chugai Pharmaceutical, and Astellas Pharma outside of this study. Y. Togashi received research grants from Ono Pharmaceutical, Bristol-Myers Squibb, Daiichi-Sankyo, Janssen Pharmaceutical, AstraZeneca, KOTAI, and KORTUC outside of this study. The other authors declare that they have no research support relevant to financial competing interests.
Figures





References
-
- Nagasaki J., et al. , PD-1 blockade therapy promotes infiltration of tumor-attacking exhausted T cell clonotypes. Cell Rep. 38, 110331 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- 22K1945904/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23KK0419/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23KK0419/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23KK0419/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21H05051/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21H02772/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22K0824/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22K20824/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22K15472/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21cm0106383/Japan Agency for Medical Research and Development (AMED)
- 21cm0106383/Japan Agency for Medical Research and Development (AMED)
- 22ama221303h0001/Japan Agency for Medical Research and Development (AMED)
- 22ama221303h0001/Japan Agency for Medical Research and Development (AMED)
- JP22ck0106775h0001/Japan Agency for Medical Research and Development (AMED)
- 23ck0106724/Japan Agency for Medical Research and Development (AMED)
- 22gm1810002s0101/Japan Agency for Medical Research and Development (AMED)
- JP22zf0127009/Japan Agency for Medical Research and Development (AMED)
- JPMJFR2049/MEXT | Japan Science and Technology Agency (JST)
- JPMJAX2321/MEXT | Japan Science and Technology Agency (JST)
- 2023-A-05/National Cancer Center Research and Development Fund
- N/A/Princess Takamatsu Cancer Research Fund
- N/A/Takeda Science Foundation (TSF)
- N/A/Naito Foundation ()
- N/A/Naito Foundation ()
- N/A/Astellas Foundation for Research on Metabolic Disorders
- N/A/MSD Life Science Foundation, Public Interest Incorporated Foundation (SD Life Science Foundation)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

