Transcriptomic analysis of BM-MSCs identified EGR1 as a transcription factor to fully exploit their therapeutic potential
- PMID: 39168411
- PMCID: PMC11480207
- DOI: 10.1016/j.bbamcr.2024.119818
Transcriptomic analysis of BM-MSCs identified EGR1 as a transcription factor to fully exploit their therapeutic potential
Abstract
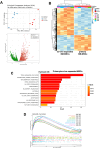
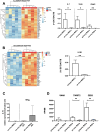
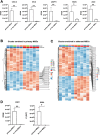
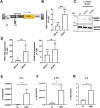
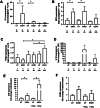
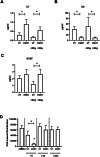
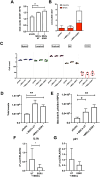
Bone marrow-mesenchymal stromal cells (BM-MSCs) are key components of the BM niche, where they regulate hematopoietic stem progenitor cell (HSPC) homeostasis by direct contact and secreting soluble factors. BM-MSCs also protect the BM niche from excessive inflammation by releasing anti-inflammatory factors and modulating immune cell activity. Thanks to these properties, BM-MSCs were successfully employed in pre-clinical HSPC transplantation models, increasing the rate of HSPC engraftment, accelerating the hematological reconstitution, and reducing the risk of graft failure. However, their clinical use requires extensive in vitro expansion, potentially altering their biological and functional properties. In this work, we analyzed the transcriptomic profile of human BM-MSCs sorted as CD45-, CD105+, CD73+, and CD90+ cells from the BM aspirates of heathy-donors and corresponding ex-vivo expanded BM-MSCs. We found the expression of immune and inflammatory genes downregulated upon cell culture and selected the transcription factor EGR1 to restore the MSC properties. We overexpressed EGR1 in BM-MSCs and performed in vitro tests to study the functional properties of EGR1-overexpressing BM-MSCs. We concluded that EGR1 increased the MSC response to inflammatory stimuli and immune cell control and potentiated the MSC hematopoietic supportive activity in co-culture assay, suggesting that the EGR1-based reprogramming may improve the BM-MSC clinical use.
Keywords: Anti-inflammatory response; Bone marrow-mesenchymal stromal cells (BM-MSCs); Hematopoietic support; MSC reprogramming; Transcriptomic analysis.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare no conflicts of interest.
Figures

Similar articles
-
Bone marrow stromal cells from β-thalassemia patients have impaired hematopoietic supportive capacity.J Clin Invest. 2019 Feb 25;129(4):1566-1580. doi: 10.1172/JCI123191. eCollection 2019 Feb 25. J Clin Invest. 2019. PMID: 30830876 Free PMC article.
-
G-CSF treatment of healthy pediatric donors affects their hematopoietic microenvironment through changes in bone marrow plasma cytokines and stromal cells.Cytokine. 2021 Mar;139:155407. doi: 10.1016/j.cyto.2020.155407. Epub 2020 Dec 28. Cytokine. 2021. PMID: 33383380
-
Direct Comparison of Wharton's Jelly and Bone Marrow-Derived Mesenchymal Stromal Cells to Enhance Engraftment of Cord Blood CD34(+) Transplants.Stem Cells Dev. 2015 Nov 15;24(22):2649-59. doi: 10.1089/scd.2015.0138. Epub 2015 Oct 8. Stem Cells Dev. 2015. PMID: 26414086 Free PMC article.
-
Mesenchymal Stromal Cells: Role in the BM Niche and in the Support of Hematopoietic Stem Cell Transplantation.Hemasphere. 2018 Nov 16;2(6):e151. doi: 10.1097/HS9.0000000000000151. eCollection 2018 Dec. Hemasphere. 2018. PMID: 31723790 Free PMC article. Review.
-
Role of ex vivo Expanded Mesenchymal Stromal Cells in Determining Hematopoietic Stem Cell Transplantation Outcome.Front Cell Dev Biol. 2021 May 4;9:663316. doi: 10.3389/fcell.2021.663316. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34017834 Free PMC article. Review.
Cited by
-
Bone marrow-derived mesenchymal stromal cells in necrotizing enterocolitis treatment: a narrative review.Front Pediatr. 2025 Jul 31;13:1624236. doi: 10.3389/fped.2025.1624236. eCollection 2025. Front Pediatr. 2025. PMID: 40822681 Free PMC article. Review.
References
-
- Michigami T., Shimizu N., Williams P.J., Niewolna M., Dallas S.L., Mundy G.R., Yoneda T. Cell-cell contact between marrow stromal cells and myeloma cells via VCAM-1 and alpha(4)beta(1)-integrin enhances production of osteoclast-stimulating activity. Blood. 2000;96:1953–1960. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

