Progesterone receptor is constitutively expressed in induced Pluripotent Stem Cells (iPSCs)
- PMID: 39168923
- PMCID: PMC11554879
- DOI: 10.1007/s12015-024-10776-6
Progesterone receptor is constitutively expressed in induced Pluripotent Stem Cells (iPSCs)
Abstract
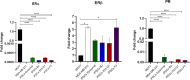
Induced Pluripotent Stem Cells (iPSCs) are nowadays a common starting point for wide-ranging applications including 3D disease modeling (i.e. organoids) and in future regenerative medicine. Physiological processes like homeostasis, cell differentiation, development and reproduction are tightly regulated by hormones through binding to their transmembrane or nuclear receptors of target cells. Considering their pleiotropic effect, take into account also their expression in an iPSCs-based disease modeling would better recapitulate the molecular events leading to 3D organoid development and disease study. Here we reported the expression pattern of estrogen receptor (ERα) and progesterone receptor (PR) in four different iPSCs, obtained from CD34 + progenitor cells and skin fibroblasts with four different methods. Expression of ERα and PR mRNA were significantly downregulated in iPSCs as well as fibroblasts compared to MCF7 positive control. Immunofluorescence (IF) staining detected only the expression of PR protein in all the different iPSCs cell lines, while ERα was not detectable. By flow cytometry analysis we observed that the ~ 65% of the total population of iPSCs cells expressed only PR, with 100% fold increase compared to HSPCs and fibroblasts, while ERα was not expressed. Our results collectively demonstrated for the first time that the reprogramming of somatic cells into iPSCs leads to the expression of PR receptor.
Keywords: CD34; Differentiation; Estrogen receptor; Fibroblasts; Induced pluripotent stem cells (iPSCs); Progesterone receptor.
© 2024. The Author(s).
Conflict of interest statement
Figures







References
-
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell2007, 131, 10.1016/j.cell.2007.11.019. - PubMed
-
- Giallongo, S.; Rehakova, D.; Raffaele, M.; Lo Re, O.; Koutna, I.; Vinciguerra, M. Redox and Epigenetics in Human Pluripotent Stem Cells Differentiation. Antioxidants Redox Signal. 2021, 34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

