Limb reduction in an Esco2 cohesinopathy mouse model is mediated by p53-dependent apoptosis and vascular disruption
- PMID: 39168984
- PMCID: PMC11339411
- DOI: 10.1038/s41467-024-51328-3
Limb reduction in an Esco2 cohesinopathy mouse model is mediated by p53-dependent apoptosis and vascular disruption
Abstract
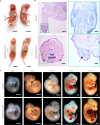
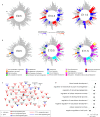
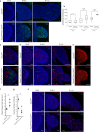
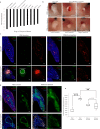
Roberts syndrome (RBS) is an autosomal recessive disorder with profound growth deficiency and limb reduction caused by ESCO2 loss-of-function variants. Here, we elucidate the pathogenesis of limb reduction in an Esco2fl/fl;Prrx1-CreTg/0 mouse model using bulk- and single-cell-RNA-seq and gene co-expression network analyses during embryogenesis. Our results reveal morphological and vascular defects culminating in hemorrhage of mutant limbs at E12.5. Underlying this abnormal developmental progression is a pre-apoptotic, mesenchymal cell population specific to mutant limb buds enriched for p53-related signaling beginning at E9.5. We then characterize these p53-related processes of cell cycle arrest, DNA damage, cell death, and the inflammatory leukotriene signaling pathway in vivo. In utero treatment with pifithrin-α, a p53 inhibitor, rescued the hemorrhage in mutant limbs. Lastly, significant enrichments were identified among genes associated with RBS, thalidomide embryopathy, and other genetic limb reduction disorders, suggesting a common vascular etiology among these conditions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R01 DE029322/DE/NIDCR NIH HHS/United States
- S10OD026880/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R03DE026814/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 AG068030/AG/NIA NIH HHS/United States
- R03 DE026814/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

