Tumor-derived RHOA mutants interact with effectors in the GDP-bound state
- PMID: 39169042
- PMCID: PMC11339415
- DOI: 10.1038/s41467-024-51445-z
Tumor-derived RHOA mutants interact with effectors in the GDP-bound state
Abstract
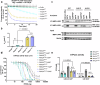
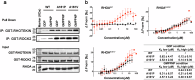
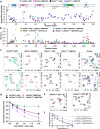
RHOA mutations are found at diverse residues in various cancer types, implying mutation- and cell-specific mechanisms of tumorigenesis. Here, we focus on the underlying mechanisms of two gain-of-function RHOA mutations, A161P and A161V, identified in adult T-cell leukemia/lymphoma. We find that RHOAA161P and RHOAA161V are both fast-cycling mutants with increased guanine nucleotide dissociation/association rates compared with RHOAWT and show reduced GTP-hydrolysis activity. Crystal structures reveal an altered nucleotide association in RHOAA161P and an open nucleotide pocket in RHOAA161V. Both mutations perturb the dynamic properties of RHOA switch regions and shift the conformational landscape important for RHOA activity, as shown by 31P NMR and molecular dynamics simulations. Interestingly, RHOAA161P and RHOAA161V can interact with effectors in the GDP-bound state. 1H-15N HSQC NMR spectra support the existence of an active population in RHOAA161V-GDP. The distinct interaction mechanisms resulting from the mutations likely favor an RHOAWT-like "ON" conformation, endowing GDP-bound state effector binding activity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

