Replay-triggered brain-wide activation in humans
- PMID: 39169063
- PMCID: PMC11339350
- DOI: 10.1038/s41467-024-51582-5
Replay-triggered brain-wide activation in humans
Abstract
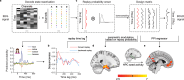
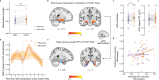
The consolidation of discrete experiences into a coherent narrative shapes the cognitive map, providing structured mental representations of our experiences. In this process, past memories are reactivated and replayed in sequence, fostering hippocampal-cortical dialogue. However, brain-wide engagement coinciding with sequential reactivation (or replay) of memories remains largely unexplored. In this study, employing simultaneous EEG-fMRI, we capture both the spatial and temporal dynamics of memory replay. We find that during mental simulation, past memories are replayed in fast sequences as detected via EEG. These transient replay events are associated with heightened fMRI activity in the hippocampus and medial prefrontal cortex. Replay occurrence strengthens functional connectivity between the hippocampus and the default mode network, a set of brain regions key to representing the cognitive map. On the other hand, when subjects are at rest following learning, memory reactivation of task-related items is stronger than that of pre-learning rest, and is also associated with heightened hippocampal activation and augmented hippocampal connectivity to the entorhinal cortex. Together, our findings highlight a distributed, brain-wide engagement associated with transient memory reactivation and its sequential replay.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Wilson, M. A. & McNaughton, B. L. Reactivation of hippocampal ensemble memories during sleep. Science265, 676–679 (1994). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

