Identification of leukemia stem cell subsets with distinct transcriptional, epigenetic and functional properties
- PMID: 39169113
- PMCID: PMC11436360
- DOI: 10.1038/s41375-024-02358-9
Identification of leukemia stem cell subsets with distinct transcriptional, epigenetic and functional properties
Abstract
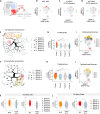
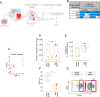
The leukemia stem cell (LSC) compartment is a complex reservoir fueling disease progression in acute myeloid leukemia (AML). The existence of heterogeneity within this compartment is well documented but prior studies have focused on genetic heterogeneity without being able to address functional heterogeneity. Understanding this heterogeneity is critical for the informed design of therapies targeting LSC, but has been hampered by LSC scarcity and the lack of reliable cell surface markers for viable LSC isolation. To overcome these challenges, we turned to the patient-derived OCI-AML22 cell model. This model includes functionally, transcriptionally and epigenetically characterized LSC broadly representative of LSC found in primary AML samples. Focusing on the pool of LSC, we used an integrated approach combining xenograft assays with single-cell analysis to identify two LSC subtypes with distinct transcriptional, epigenetic and functional properties. These LSC subtypes differed in depth of quiescence, differentiation potential, repopulation capacity, sensitivity to chemotherapy and could be isolated based on CD112 expression. A majority of AML patient samples had transcriptional signatures reflective of either LSC subtype, and some even showed coexistence within an individual sample. This work provides a framework for investigating the LSC compartment and designing combinatorial therapeutic strategies in AML.
© 2024. The Author(s).
Conflict of interest statement
JED reports receiving a commercial research grant from Celgene/BMS, and JED and JCYW have patents licensed to Trillium Therapeutics/Pfizer.
Figures




References
-
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–93. - PubMed
-
- Ng SWK, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–7. - PubMed
-
- van Rhenen A, Feller N, Kelder A, Westra AH, Rombouts E, Zweegman S, et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res. 2005;11:6520–7. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

