Novel immunotherapeutics against LGR5 to target multiple cancer types
- PMID: 39169164
- PMCID: PMC11393416
- DOI: 10.1038/s44321-024-00121-2
Novel immunotherapeutics against LGR5 to target multiple cancer types
Erratum in
-
Publisher Correction: Novel immunotherapeutics against LGR5 to target multiple cancer types.EMBO Mol Med. 2024 Nov;16(11):3026. doi: 10.1038/s44321-024-00139-6. EMBO Mol Med. 2024. PMID: 39322863 Free PMC article.
Abstract
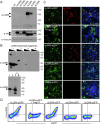
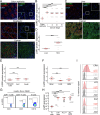
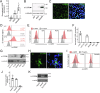
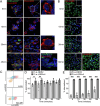
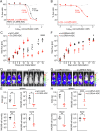
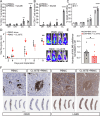
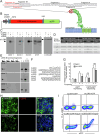
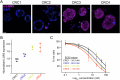
We have developed and validated a highly specific, versatile antibody to the extracellular domain of human LGR5 (α-LGR5). α-LGR5 detects LGR5 overexpression in >90% of colorectal cancer (CRC), hepatocellular carcinoma (HCC) and pre-B-ALL tumour cells and was used to generate an Antibody-Drug Conjugate (α-LGR5-ADC), Bispecific T-cell Engager (α-LGR5-BiTE) and Chimeric Antigen Receptor (α-LGR5-CAR). α-LGR5-ADC was the most effective modality for targeting LGR5+ cancer cells in vitro and demonstrated potent anti-tumour efficacy in a murine model of human NALM6 pre-B-ALL driving tumour attrition to less than 1% of control treatment. α-LGR5-BiTE treatment was less effective in the pre-B-ALL cancer model yet promoted a twofold reduction in tumour burden. α-LGR5-CAR-T cells also showed specific and potent LGR5+ cancer cell killing in vitro and effective tumour targeting with a fourfold decrease in pre-B-ALL tumour burden relative to controls. Taken together, we show that α-LGR5 can not only be used as a research tool and a biomarker but also provides a versatile building block for a highly effective immune therapeutic portfolio targeting a range of LGR5-expressing cancer cells.
Keywords: ADC; BiTE; CAR; Cancer Immunotherapeutics; LGR5.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. The LGR5 antibodies in the manuscript and all commercial use fall under patent filing PCT/GB2023/050512, inventors—Maike de la Roche and Marc de la Roche.
Figures

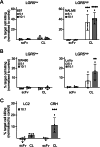
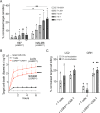
References
MeSH terms
Substances
Grants and funding
- 3903171/Pathological Society of Great Britain and Ireland
- 22257/CRUK_/Cancer Research UK/United Kingdom
- RG94521/UKRI | Medical Research Council (MRC)
- 107609/Z/15/Z/Wellcome Trust (WT)
- RCCCSF-May23/100001/Cancer Research UK (CRUK)
- 227432/Z/23/Z/Wellcome Trust (WT)
- 203151/Z/16/Z/Wellcome Trust (WT)
- 222062/Z/20/Z/Wellcome Trust (WT)
- C52489/A19924/Cancer Research UK (CRUK)
- WT_/Wellcome Trust/United Kingdom
- C67279/A27957/Cancer Research UK (CRUK)
- A22257/Cancer Research UK (CRUK)
- C18873/A26813/Cancer Research UK (CRUK)
- C63389/A30462/Cancer Research UK (CRUK)
LinkOut - more resources
Full Text Sources

