Genetic architecture of epigenetic cortical clock age in brain tissue from older individuals: alterations in CD46 and other loci
- PMID: 39169872
- PMCID: PMC11346548
- DOI: 10.1080/15592294.2024.2392050
Genetic architecture of epigenetic cortical clock age in brain tissue from older individuals: alterations in CD46 and other loci
Abstract
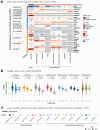
The cortical epigenetic clock was developed in brain tissue as a biomarker of brain aging. As one way to identify mechanisms underlying aging, we conducted a GWAS of cortical age. We leveraged postmortem cortex tissue and genotyping array data from 694 participants of the Rush Memory and Aging Project and Religious Orders Study (ROSMAP; 11000,000 SNPs), and meta-analysed ROSMAP with 522 participants of Brains for Dementia Research (5,000,000 overlapping SNPs). We confirmed results using eQTL (cortical bulk and single nucleus gene expression), cortical protein levels (ROSMAP), and phenome-wide association studies (clinical/neuropathologic phenotypes, ROSMAP). In the meta-analysis, the strongest association was rs4244620 (p = 1.29 × 10-7), which also exhibited FDR-significant cis-eQTL effects for CD46 in bulk and single nucleus (microglia, astrocyte, oligodendrocyte, neuron) cortical gene expression. Additionally, rs4244620 was nominally associated with lower cognition, faster slopes of cognitive decline, and greater Parkinsonian signs (n ~ 1700 ROSMAP with SNP/phenotypic data; all p ≤ 0.04). In ROSMAP alone, the top SNP was rs4721030 (p = 8.64 × 10-8) annotated to TMEM106B and THSD7A. Further, in ROSMAP (n = 849), TMEM106B and THSD7A protein levels in cortex were related to many phenotypes, including greater AD pathology and lower cognition (all p ≤ 0.0007). Overall, we identified converging evidence of CD46 and possibly TMEM106B/THSD7A for potential roles in cortical epigenetic clock age.
Keywords: Aging; brain; dementia; epigenetics.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
