Single-cell RNA and T-cell receptor sequencing unveil mycosis fungoides heterogeneity and a possible gene signature
- PMID: 39169943
- PMCID: PMC11337020
- DOI: 10.3389/fonc.2024.1408614
Single-cell RNA and T-cell receptor sequencing unveil mycosis fungoides heterogeneity and a possible gene signature
Abstract
Background: Mycosis fungoides (MF) is the most common subtype of cutaneous T-cell lymphoma (CTCL). Comprehensive analysis of MF cells in situ and ex vivo is complicated by the fact that is challenging to distinguish malignant from reactive T cells with certainty.
Methods: To overcome this limitation, we performed combined single-cell RNA (scRNAseq) and T-cell receptor TCR sequencing (scTCRseq) of skin lesions of cutaneous MF lesions from 12 patients. A sufficient quantity of living T cells was obtained from 9 patients, but 2 had to be excluded due to unclear diagnoses (coexisting CLL or revision to a fixed toxic drug eruption).
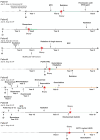
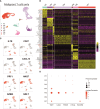
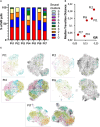
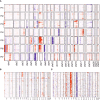
Results: From the remaining patients we established single-cell mRNA expression profiles and the corresponding TCR repertoire of 18,630 T cells. TCR clonality unequivocally identified 13,592 malignant T cells. Reactive T cells of all patients clustered together, while malignant cells of each patient formed a unique cluster expressing genes typical of naive/memory, such as CD27, CCR7 and IL7R, or cytotoxic T cells, e.g., GZMA, NKG7 and GNLY. Genes encoding classic CTCL markers were not detected in all clusters, consistent with the fact that mRNA expression does not correlate linearly with protein expression. Nevertheless, we successfully pinpointed distinctive gene signatures differentiating reactive malignant from malignant T cells: keratins (KRT81, KRT86), galectins (LGALS1, LGALS3) and S100 genes (S100A4, S100A6) being overexpressed in malignant cells.
Conclusions: Combined scRNAseq and scTCRseq not only allows unambiguous identification of MF cells, but also revealed marked heterogeneity between and within patients with unexpected functional phenotypes. While the correlation between mRNA and protein abundance was limited with respect to established MF markers, we were able to identify a single-cell gene expression signature that distinguishes malignant from reactive T cells.
Keywords: CTCL; TCR sequencing; gene signature; heterogeneity; malignant T cells; single-cell RNA sequencing.
Copyright © 2024 Srinivas, Peiffer, Horny, Lei, Buus, Kubat, Luo, Yin, Spassova, Sucker, Farahpour, Kehrmann, Ugurel, Livingstone, Gambichler, Ødum and Becker.
Conflict of interest statement
SU declares research support from Bristol Myers Squibb and Merck Serono; speakers and advisory board honoraria from Bristol Myers Squibb, Merck Sharp and Novartis, and travel support from Bristol Myers Squibb, Merck Sharp & Dohme and Pierre Fabre. EL reports personal fees, honoraria, and support for attending meetings or travel grants from Bristol Myers Squibb, Medac, Novartis, Pierre Fabre, and Sun Pharma, and honoraria from MSD, Recordati, and Sanofi. EL participated on a drug safety monitoring or advisory board for Bristol Myers Squibb, Novartis, Sanofi, and Sun Pharma. TG has received speakers and/or advisory board honoraria from BMS, Sano-fi-Genzyme, MSD, Novartis Pharma, Roche, Abbvie, Almirall, Janssen, Lilly, Pfizer, Pierre Fabre, Merck-Serono, outside the submitted work. JB is receiving speaker’s bureau honoraria from Amgen, Pfizer, Recordati and Sanofi, and is a paid consultant/advisory board member/DSMB member for Almirall, Boehringer Ingelheim, InProTher, ICON, Merck Serono, Pfizer, 4SC and Sanofi/Regeneron. His group receives research grants from Bristol Myers Squibb, Merck Serono, HTG, IQVIA and Alcedis GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

