Exploring Indonesian actinomycete extracts for anti-tubercular compounds: Integrating inhibition assessment, genomic analysis, and prediction of its target by molecular docking
- PMID: 39170210
- PMCID: PMC11336835
- DOI: 10.1016/j.heliyon.2024.e35648
Exploring Indonesian actinomycete extracts for anti-tubercular compounds: Integrating inhibition assessment, genomic analysis, and prediction of its target by molecular docking
Abstract
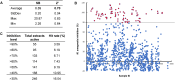
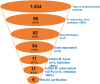
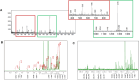
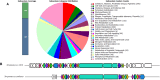
Tuberculosis (TB) is the foremost cause of infectious fatality globally. The primary global challenge in combatting TB lies in addressing the emergence of drug-resistant variants of the disease. However, the number of newly approved agents for treating TB has remained remarkably low over recent decades. Hence, research endeavors for discovering novel anti-TB agents are always needed. In the present study, we screened over 1,500 culture extracts from actinomycetes isolated in Indonesia for their inhibitory activity against Mycobacterium smegmatis used as a surrogate in the primary screening. The initial screening yielded approximately 6.2 % hit extracts, with a selection criterion of >80 % growth inhibition. The confirmed hit extracts were subsequently subjected to growth inhibition assay against Mycobacterium bovis and Mycobacterium tuberculosis. Approximately 20 % of the hit extracts that showed growth inhibition also exhibited efficacy against M. bovis BCG and M. tuberculosis H37Rv pathogenic strain. An active compound was successfully purified from a large-scale culture of the most potent representative extract by high-performance liquid chromatography and thin-layer chromatography. The structure of the active compound was elucidated by mass spectrometry and nuclear magnetic resonance. This compound displayed structural similarities to actinomycin group and exhibited robust inhibition, with IC50 values of 0.74, 0.02, and 0.07 μg/mL against M. smegmatis, M. bovis, and M. tuberculosis, respectively. The Actinomycetes strain A612, which produced the active compound, was taxonomically classified by phylogenetic analysis of 16s rRNA gene and whole genome sequencing data as Streptomyces parvus. Computational genome analysis utilizing anti-SMASH 7.0 unveiled that S. parvus A612 strain harbors 40 biosynthetic gene clusters with the potential to produce 16 known (with >70 % similarity) and 24 unknown compounds. A non-ribosomal peptide synthesis (NRPS) gene cluster associated with actinomycin D biosynthesis was also identified, boasting an 85 % similarity. Molecular docking analysis of actinomycin D and 21 potential M. tuberculosis targets revealed possible interactions with multiple targets. The purified active compound inhibited recombinant M. tuberculosis shikimate kinase (MtSK), which validated the results obtained from the docking analysis.
Keywords: Actinomycetes; Active compound; Docking study; Extracts; Screening; Shikimate kinase; Tuberculosis; Whole genome.
© 2024 The Author(s).
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Arif Nurkanto and Tomoyoshi Nozaki reports financial support, administrative support, article publishing charges, equipment, drugs, or supplies, and travel were provided by Rispro 10.13039/501100014538LPDP Indonesia, 10.13039/501100009037Science and Technology Research Partnership for Sustainable Development (10.13039/501100009037SATREPS) from the 10.13039/100009619Japan Agency for Medical Research and Development (10.13039/100009619AMED) and 10.13039/501100004532Japan International Cooperation Agency (10.13039/501100004532JICA). If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
A New Screen for Tuberculosis Drug Candidates Utilizing a Luciferase-Expressing Recombinant Mycobacterium bovis Bacillus Calmette-Guéren.PLoS One. 2015 Nov 16;10(11):e0141658. doi: 10.1371/journal.pone.0141658. eCollection 2015. PLoS One. 2015. PMID: 26571296 Free PMC article.
-
Identification of novel antimicrobial compounds targeting Mycobacterium tuberculosis shikimate kinase using in silico hierarchical structure-based drug screening.Tuberculosis (Edinb). 2023 Jul;141:102362. doi: 10.1016/j.tube.2023.102362. Epub 2023 Jun 8. Tuberculosis (Edinb). 2023. PMID: 37311288
-
A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity.Appl Microbiol Biotechnol. 2012 Aug;95(4):919-27. doi: 10.1007/s00253-012-4079-z. Epub 2012 Apr 28. Appl Microbiol Biotechnol. 2012. PMID: 22543353
-
[Development of antituberculous drugs: current status and future prospects].Kekkaku. 2006 Dec;81(12):753-74. Kekkaku. 2006. PMID: 17240921 Review. Japanese.
-
Shikimate Kinase Inhibitors: An Update on Promising Strategy against Mycobacterium tuberculosis.Curr Drug Targets. 2023;24(5):388-405. doi: 10.2174/1389450124666230208102645. Curr Drug Targets. 2023. PMID: 36752299 Review.
Cited by
-
Innovative Strategies for Combating Multidrug-Resistant Tuberculosis: Advances in Drug Delivery Systems and Treatment.Microorganisms. 2025 Mar 24;13(4):722. doi: 10.3390/microorganisms13040722. Microorganisms. 2025. PMID: 40284559 Free PMC article. Review.
References
-
- Hershkovitz I., Donoghue H.D., Minnikin D.E., Besra G.S., Lee O.Y.C., Gernaey A.M., Galili E., Eshed V., Greenblatt C.L., Lemma E., Bar-Gal G.K., Spigelman M. Detection and molecular characterization of 9000-year-old Mycobacterium tuberculosis from a neolithic settlement in the eastern mediterranean. PLoS One. 2008;3 doi: 10.1371/JOURNAL.PONE.0003426. - DOI - PMC - PubMed
-
- World Health Organization Global tuberculosis report 2022, Work. Heal. Saf. 2022:68. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... August 8, 2023.
-
- World Health Organization, Global tuberculosis report (2020). https://www.who.int/publications/i/item/9789240013131. (Accessed 31 August 2023).
-
- Klopper M., Warren R.M., Hayes C., van Pittius N.C.G., Streicher E.M., Müller B., Sirgel F.A., Chabula-Nxiweni M., Hoosain E., Coetzee G., van Helden P.D., Victor T.C., Trollip A.P. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 2013;19:449–455. doi: 10.3201/EID1903.120246. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

