Differential effect of asparagine and glutamine removal on three adenocarcinoma cell lines
- PMID: 39170541
- PMCID: PMC11337022
- DOI: 10.1016/j.heliyon.2024.e35789
Differential effect of asparagine and glutamine removal on three adenocarcinoma cell lines
Abstract
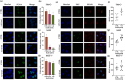
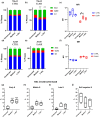
Asparagine and glutamine depletion operated by the drug Asparaginase (ASNase) has revolutionized therapy in pediatric patients affected by Acute Lymphoblastic Leukemia (ALL), bringing remissions to a remarkable 90 % of cases. However, the knowledge of the proproliferative role of asparagine in adult and solid tumors is still limited. We have here analyzed the effect of ASNase on three adenocarcinoma cell lines (A549, lung adenocarcinoma, MCF-7, breast cancer, and 786-O, kidney cancer). In contrast to MCF-7 cells, 786-O and A549 cells proved to be a relevant target for cell cycle perturbation by asparagine and glutamine shortage. Indeed, when the cell-cycle was analyzed by flow cytometry, A549 showed a canonical response to asparaginase, 786-O cells, instead, showed a reduction of the percentage of cells in the G1 phase and an increase of those in the S-phase. Despite an increased number of PCNA and RPA70 positive nuclear foci, BrdU and EdU incorporation was absent or strongly delayed in treated 786-O cells, thus indicating a readiness of replication forks unmatched by DNA synthesis. In 786-O asparagine synthetase was reduced following treatment and glutamine synthetase was totally absent. Interestingly, DNA synthesis could be recovered by adding Gln to the medium. MCF-7 cells showed no significant changes in the cell cycle phases, in DNA-bound PCNA and in total PCNA, but a significant increase in ASNS and GS mRNA and protein expression. The collected data suggest that the effect observed on 786-O cells following ASNase treatment could rely on mechanisms which differ from those well-known and described for leukemic blasts, consisting of a complete block in the G1/S transition in proliferating cells and on an increase on non-proliferative (G0) blasts.
Keywords: Adenocarcinoma; Asparaginase; Asparagine synthetase; Breast carcinoma; Cell cycle; Glutamine synthetase; Renal cell carcinoma; Solid tumors.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Maristella Maggi has patent A HIGHLY STABLE, PROTEASE-RESISTANT E. COLI ASPARAGINASE issued to University of Pavia. Claudia Scotti has patent A HIGHLY STABLE, PROTEASE-RESISTANT E. COLI ASPARAGINASE issued to University of Pavia. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Zhang J., Fan J., Venneti S., Cross J.R., Takagi T., Bhinder B., Djaballah H., Kanai M., Cheng E.H., Judkins A.R., Pawel B., Baggs J., Cherry S., Rabinowitz J.D., Thompson C.B. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol. Cell. 2014;56:205–218. doi: 10.1016/J.MOLCEL.2014.08.018. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

