Gut resistome of NSCLC patients treated with immunotherapy
- PMID: 39170692
- PMCID: PMC11335565
- DOI: 10.3389/fgene.2024.1378900
Gut resistome of NSCLC patients treated with immunotherapy
Abstract
Background: The newest method of treatment for patients with NSCLC (non-small cell lung cancer) is immunotherapy directed at the immune checkpoints PD-1 (Programmed Cell Death 1) and PD-L1 (Programmed Cell Death Ligand 1). PD-L1 is the only validated predictor factor for immunotherapy efficacy, but it is imperfect. Some patients do not benefit from immunotherapy and may develop primary or secondary resistance. This study aimed to assess the intestinal resistome composition of non-small cell lung cancer (NSCLC) patients treated with immune checkpoint inhibitors in the context of clinical features and potentially new prediction factors for assessing immunotherapy efficacy.
Methods: The study included 30 advanced NSCLC patients, 19 (57%) men and 11 (33%) women treated with first- or second-line immunotherapy (nivolumab, pembrolizumab or atezolizumab). We evaluated the patient's gut resistome composition using the high sensitivity of targeted metagenomics.
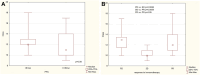
Results: Studies have shown that resistome richness is associated with clinical and demographic factors of NSCLC patients treated with immunotherapy. Smoking seems to be associated with an increased abundance of macrolides, lincosamides, streptogramins and vancomycin core resistome. The resistome of patients with progression disease appears to be more abundant and diverse, with significantly higher levels of genomic markers of resistance to lincosamides (lnuC). The resistance genes lnuC, msrD, ermG, aph(6), fosA were correlated with progression-free survival or/and overall survival, thus may be considered as factors potentially impacting the disease.
Conclusion: The results indicate that the intestinal resistome of NSCLC patients with immune checkpoint inhibitors treatment differs depending on the response to immunotherapy, with several distinguished markers. Since it might impact treatment efficacy, it must be examined more deeply.
Keywords: NSCLC; immunotherapy; metagenomics; microbiome; resistome.
Copyright © 2024 Iwan, Grenda, Bomba, Bielińska, Wasyl, Kieszko, Rolska-Kopińska, Chmielewska, Krawczyk, Rybczyńska-Tkaczyk, Olejnik and Milanowski.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. 10.1093/nar/gkac920 - DOI - PMC - PubMed
-
- Bavaro D. F., Pizzutilo P., Catino A., Signorile F., Pesola F., Di Gennaro F., et al. (2021). Incidence of infections and predictors of mortality during checkpoint inhibitor immunotherapy in patients with advanced lung cancer: a retrospective cohort study. Open Forum Infect. Dis. 8 (6), ofab187. 10.1093/ofid/ofab187 - DOI - PMC - PubMed
-
- Bonin N., Doster E., Worley H., Pinnell L. J., Bravo J. E., Ferm P., et al. (2023). MEGARes and AMR++, v3.0: an updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 51, D744–D752. 10.1093/nar/gkac1047 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

