Bi-functional CpG-STAT3 decoy oligonucleotide triggers multilineage differentiation of acute myeloid leukemia in mice
- PMID: 39171140
- PMCID: PMC11338104
- DOI: 10.1016/j.omtn.2024.102268
Bi-functional CpG-STAT3 decoy oligonucleotide triggers multilineage differentiation of acute myeloid leukemia in mice
Abstract
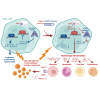
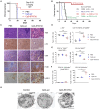
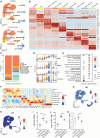
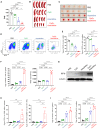
Acute myeloid leukemia (AML) cells resist differentiation stimuli despite high expression of innate immune receptors, such as Toll-like receptor 9 (TLR9). We previously demonstrated that targeting Signal Transducer and Activator of Transcription 3 (STAT3) using TLR9-targeted decoy oligodeoxynucleotide (CpG-STAT3d) increases immunogenicity of human and mouse AML cells. Here, we elucidated molecular mechanisms of inv(16) AML reprogramming driven by STAT3-inhibition/TLR9-activation in vivo. At the transcriptional levels, AML cells isolated from mice after intravenous administration of CpG-STAT3d or leukemia-targeted Stat3 silencing and TLR9 co-stimulation, displayed similar upregulation of myeloid cell differentiation (Irf8, Cebpa, Itgam) and antigen-presentation (Ciita, Il12a, B2m)-related genes with concomitant reduction of leukemia-promoting Runx1. Single-cell transcriptomics revealed that CpG-STAT3d induced multilineage differentiation of AML cells into monocytes/macrophages, erythroblastic and B cell subsets. As shown by an inducible Irf8 silencing in vivo, IRF8 upregulation was critical for monocyte-macrophage differentiation of leukemic cells. TLR9-driven AML cell reprogramming was likely enabled by downregulation of STAT3-controlled methylation regulators, such as DNMT1 and DNMT3. In fact, the combination of DNA methyl transferase (DNMT) inhibition using azacitidine with CpG oligonucleotides alone mimicked CpG-STAT3d effects, resulting in AML cell differentiation, T cell activation, and systemic leukemia regression. These findings highlight immunotherapeutic potential of bi-functional oligonucleotides to unleash TLR9-driven differentiation of leukemic cells by concurrent STAT3 and/or DNMT inhibition.
Keywords: CpG deoxynucleotides; MT: Oligonucleotides: Therapies and Applications; Oligonucleotide therapeutics; STAT3; TLR9; acute myeloid leukemia; cancer immunotherapy; decoy DNA.
© 2024 The Author(s).
Conflict of interest statement
M.K. is an inventor on the patents that cover the design of CpG-STAT3d ODNs. M.K. serves on the Scientific Advisory Board of Scopus Biopharma and its subsidiary Duet Biotherapeutics with stock options and sponsored research.
Figures


References
-
- Khwaja A., Bjorkholm M., Gale R.E., Levine R.L., Jordan C.T., Ehninger G., Bloomfield C.D., Estey E., Burnett A., Cornelissen J.J., et al. Acute myeloid leukaemia. Nat. Rev. Dis. Prim. 2016;2 - PubMed
-
- Boyiadzis M., Bishop M.R., Abonour R., Anderson K.C., Ansell S.M., Avigan D., Barbarotta L., Barrett A.J., Van Besien K., Bergsagel P.L., et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of hematologic malignancies: multiple myeloma, lymphoma, and acute leukemia. J. Immunother. Cancer. 2016;4:90. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

