Data-driven biomarkers better associate with stroke motor outcomes than theory-based biomarkers
- PMID: 39171205
- PMCID: PMC11336660
- DOI: 10.1093/braincomms/fcae254
Data-driven biomarkers better associate with stroke motor outcomes than theory-based biomarkers
Abstract
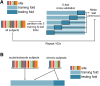
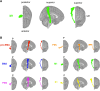
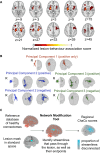
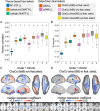
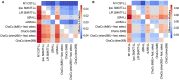
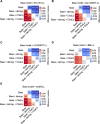
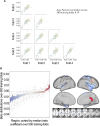
Chronic motor impairments are a leading cause of disability after stroke. Previous studies have associated motor outcomes with the degree of damage to predefined structures in the motor system, such as the corticospinal tract. However, such theory-based approaches may not take full advantage of the information contained in clinical imaging data. The present study uses data-driven approaches to model chronic motor outcomes after stroke and compares the accuracy of these associations to previously-identified theory-based biomarkers. Using a cross-validation framework, regression models were trained using lesion masks and motor outcomes data from 789 stroke patients from the Enhancing NeuroImaging Genetics through Meta Analysis (ENIGMA) Stroke Recovery Working Group. Using the explained variance metric to measure the strength of the association between chronic motor outcomes and imaging biomarkers, we compared theory-based biomarkers, like lesion load to known motor tracts, to three data-driven biomarkers: lesion load of lesion-behaviour maps, lesion load of structural networks associated with lesion-behaviour maps, and measures of regional structural disconnection. In general, data-driven biomarkers had stronger associations with chronic motor outcomes accuracy than theory-based biomarkers. Data-driven models of regional structural disconnection performed the best of all models tested (R 2 = 0.210, P < 0.001), performing significantly better than the theory-based biomarkers of lesion load of the corticospinal tract (R 2 = 0.132, P < 0.001) and of multiple descending motor tracts (R 2 = 0.180, P < 0.001). They also performed slightly, but significantly, better than other data-driven biomarkers including lesion load of lesion-behaviour maps (R 2 = 0.200, P < 0.001) and lesion load of structural networks associated with lesion-behaviour maps (R 2 = 0.167, P < 0.001). Ensemble models - combining basic demographic variables like age, sex, and time since stroke - improved the strength of associations for theory-based and data-driven biomarkers. Combining both theory-based and data-driven biomarkers with demographic variables improved predictions, and the best ensemble model achieved R 2 = 0.241, P < 0.001. Overall, these results demonstrate that out-of-sample associations between chronic motor outcomes and data-driven imaging features, particularly when lesion data is represented in terms of structural disconnection, are stronger than associations between chronic motor outcomes and theory-based biomarkers. However, combining both theory-based and data-driven models provides the most robust associations.
Keywords: imaging biomarkers; lesion-deficit associations; machine learning; stroke outcomes.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
S.C.C. serves as a consultant for Abbvie, Constant Therapeutics, BrainQ, Myomo, MicroTransponder, Neurolutions, Panaxium, NeuExcell, Elevian, Helius, Omniscient, Brainsgate, Nervgen, Battelle, and TRCare. B.H. has a clinical partnership with Fourier Intelligence. N.J.S. is an inventor for a patent US 10,071,015 B2. C.J. W. is a consultant for Microtransponder, BrainQ, and MedRhythm. G.F.W. sits on Advisory Boards for Myomo and Neuro-innovators.
Figures

Update of
-
Data-driven biomarkers outperform theory-based biomarkers in predicting stroke motor outcomes.bioRxiv [Preprint]. 2023 Sep 1:2023.06.19.545638. doi: 10.1101/2023.06.19.545638. bioRxiv. 2023. Update in: Brain Commun. 2024 Jul 31;6(4):fcae254. doi: 10.1093/braincomms/fcae254. PMID: 37693419 Free PMC article. Updated. Preprint.
References
-
- Kelly-Hayes M, Beiser A, Kase CS, Scaramucci A, D’Agostino RB, Wolf PA. The influence of gender and age on disability following ischemic stroke: The Framingham study. J Stroke Cerebrovasc Dis. 2003;12(3):119–126. - PubMed
Grants and funding
- R00 HD091375/HD/NICHD NIH HHS/United States
- S10 RR028821/RR/NCRR NIH HHS/United States
- R01 NS115845/NS/NINDS NIH HHS/United States
- R01 HD095457/HD/NICHD NIH HHS/United States
- U01 NS120910/NS/NINDS NIH HHS/United States
- R01 MH116147/MH/NIMH NIH HHS/United States
- R01 HD062744/HD/NICHD NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- R01 NS076348/NS/NINDS NIH HHS/United States
- U01 NS086872/NS/NINDS NIH HHS/United States
- RF1 MH123163/MH/NIMH NIH HHS/United States
- R41 HD104296/HD/NICHD NIH HHS/United States
- R01 HD065438/HD/NICHD NIH HHS/United States
- T32 GM108540/GM/NIGMS NIH HHS/United States
- UH3 NS121565/NS/NINDS NIH HHS/United States
- R01 MH121246/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 HD094731/HD/NICHD NIH HHS/United States
- P20 GM109040/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
