The molecular basis of lactase persistence: Linking genetics and epigenetics
- PMID: 39171584
- PMCID: PMC12336946
- DOI: 10.1111/ahg.12575
The molecular basis of lactase persistence: Linking genetics and epigenetics
Abstract
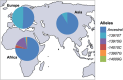
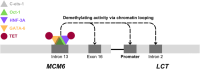
Lactase persistence (LP) - the genetic trait that determines the continued expression of the enzyme lactase into adulthood - has undergone recent, rapid positive selection since the advent of animal domestication and dairying in some human populations. While underlying evolutionary explanations have been widely posited and studied, the molecular basis of LP remains less so. This review considers the genetic and epigenetic bases of LP. Multiple single-nucleotide polymorphisms (SNPs) in an LCT enhancer in intron 13 of the neighbouring MCM6 gene are associated with LP. These SNPs alter binding of transcription factors (TFs) and likely prevent age-related increases in methylation in the enhancer, maintaining LCT expression into adulthood to cause LP. However, the complex relationship between the genetics and epigenetics of LP is not fully characterised, and the mode of action of methylation quantitative trait loci (meQTLs) (SNPs affecting methylation) generally remains poorly understood. Here, we examine published LP data to propose a model describing how methylation in the LCT enhancer is prevented in LP adults. We argue that this occurs through altered binding of the TF Oct-1 (encoded by the gene POU2F1) and neighbouring TFs GATA-6 (GATA6), HNF-3A (FOXA1) and c-Ets1 (ETS1) acting in concert. We therefore suggest a plausible new model for LCT downregulation in the context of LP, with wider relevance for future work on the mechanisms of other meQTLs.
Keywords: epigenetics; gene expression; lactase persistence; methylation; multiple alleles; transcription factors.
© 2024 The Author(s). Annals of Human Genetics published by University College London (UCL) and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Banovich, N. E. , Lan, X. , McVicker, G. , van de Geijn, B. , Degner, J. F. , Blischak, J. D. , Roux, J. , Pritchard, J. K. , & Gilad, Y. (2014). Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genetics, 10(9), Article e1004663. 10.1371/journal.pgen.1004663 - DOI - PMC - PubMed
-
- Bersaglieri, T. , Sabeti, P. C. , Patterson, N. , Vanderploeg, T. , Schaffner, S. F. , Drake, J. A. , Rhodes, M. , Reich, D. E. , & Hirschhorn, J. N. (2004). Genetic signatures of strong recent positive selection at the lactase gene. The American Journal of Human Genetics, 74(6), 1111–1120. 10.1086/421051 - DOI - PMC - PubMed
-
- Bonder, M. J. , Kurilshikov, A. , Tigchelaar, E. F. , Mujagic, Z. , Imhann, F. , Vila, A. V. , Deelen, P. , Vatanen, T. , Schirmer, M. , Smeekens, S. P. , Zhernakova, D. V. , Jankipersadsing, S. A. , Jaeger, M. , Oosting, M. , Cenit, M. C. , Masclee, A. A. , Swertz, M. A. , Li, Y. , Kumar, V. , … Zhernakova, A. (2016). The effect of host genetics on the gut microbiome. Nature Genetics, 48(11), 1407–1412. 10.1038/ng.3663 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

