Genomic implications of the repeated shift to self-fertilization across a species' geographic distribution
- PMID: 39171640
- PMCID: PMC11700596
- DOI: 10.1093/jhered/esae046
Genomic implications of the repeated shift to self-fertilization across a species' geographic distribution
Abstract
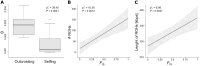
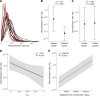
The ability to self-fertilize often varies among closely related hermaphroditic plant species, though, variation can also exist within species. In the North American Arabidopsis lyrata, the shift from self-incompatibility (SI) to selfing established in multiple regions independently, mostly since recent postglacial range expansion. This has made the species an ideal model for the investigation of the genomic basis of the breakdown of SI and its population genetic consequences. By comparing nearby selfing and outcrossing populations across the entire species' geographic distribution, we investigated variation at the self-incompatibility (S-)locus and across the genome. Furthermore, a diallel crossing experiment on one mixed-mating population was performed to gain insight into the inheritance of mating system variation. We confirmed that the breakdown of SI had evolved in several S-locus backgrounds. The diallel suggested the involvement of biparental contributions with dominance relations. Though, the population-level genome-wide association study did not single out clear-cut candidate genes but several regions with one near the S-locus. On the implication side, selfing as compared to outcrossing populations had less than half of the genomic diversity, while the number and length of runs of homozygosity (ROHs) scaled with the degree of inbreeding. Selfing populations with a history of long expansion had the longest ROHs. The results highlight that mating system shift to selfing, its genetic underpinning and the likely negative genomic consequences for evolutionary potential can be strongly interlinked with past range dynamics.
Keywords: inbreeding; mating system; recombination; runs of homozygosity; self-incompatibility locus.
© The American Genetic Association. 2024.
Figures


References
-
- Abhinandan K, Sankaranarayanan S, Macgregor S, Goring DR, Samuel MA.. Cell–cell signaling during the Brassicaceae self-incompatibility response. Trends Plant Sci. 2022:27:472–487. - PubMed
-
- Bachmann JA, Tedder A, Laenen B, Fracassetti M, Désamoré A, Lafon‐Placette C, Steige KA, Callot C, Marande W, Neuffer B, et al. Genetic basis and timing of a major mating system shift in Capsella. New Phytol. 2019:224:505–517. - PubMed
-
- Bates D, Mächler M, Bolker B, Walker S.. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015:67:1–48.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

