An approach to analyze spatiotemporal patterns of gene expression at single-cell resolution in Candida albicans-infected mouse tongues
- PMID: 39171917
- PMCID: PMC11423565
- DOI: 10.1128/msphere.00282-24
An approach to analyze spatiotemporal patterns of gene expression at single-cell resolution in Candida albicans-infected mouse tongues
Abstract
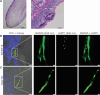
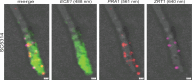
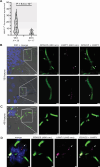
Microbial gene expression measurements derived from infected organs are invaluable to understand pathogenesis. However, current methods are limited to "bulk" analyses that neglect microbial cell heterogeneity and the lesion's spatial architecture. Here, we report the use of hybridization chain reaction RNA fluorescence in situ hybridization (HCR RNA-FISH) to visualize and quantify Candida albicans transcripts at single-cell resolution in tongues of infected mice. The method is compatible with fixed-frozen and formalin-fixed paraffin-embedded tissues. We document cell-to-cell variation and intriguing spatiotemporal expression patterns for C. albicans mRNAs that encode products implicated in oral candidiasis. The approach provides a spatial dimension to gene expression analyses of host-Candida interactions.
Importance: Candida albicans is a fungal pathobiont inhabiting multiple mucosal surfaces of the human body. Immunosuppression, antibiotic-induced microbial dysbiosis, or implanted medical devices can impair mucosal integrity enabling C. albicans to overgrow and disseminate, causing either mucosal diseases such as oropharyngeal candidiasis or life-threatening systemic infections. Profiling fungal genes that are expressed in the infected mucosa or in any other infected organ is paramount to understand pathogenesis. Ideally, these transcript profiling measurements should reveal the expression of any gene at the single-cell level. The resolution typically achieved with current approaches, however, limits most gene expression measurements to cell population averages. The approach described in this report provides a means to dissect fungal gene expression in infected tissues at single-cell resolution.
Keywords: Candida albicans; Candida-host interactions; Candida-infected tissue; RNA-FISH; hybridization chain reaction; microbial transcript visualization; oropharyngeal candidiasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Imaging and Quantification of mRNA Molecules at Single-Cell Resolution in the Human Fungal Pathogen Candida albicans.mSphere. 2021 Aug 25;6(4):e0041121. doi: 10.1128/mSphere.00411-21. Epub 2021 Jul 7. mSphere. 2021. PMID: 34232078 Free PMC article.
-
Mucosal Bacteria Modulate Candida albicans Virulence in Oropharyngeal Candidiasis.mBio. 2021 Aug 31;12(4):e0193721. doi: 10.1128/mBio.01937-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399623 Free PMC article.
-
Identification of Candida albicans regulatory genes governing mucosal infection.Cell Microbiol. 2018 Aug;20(8):e12841. doi: 10.1111/cmi.12841. Epub 2018 Apr 18. Cell Microbiol. 2018. PMID: 29575428
-
Perspective on receptor-associated immune response to Candida albicans single and mixed infections: Implications for therapeutics in oropharyngeal candidiasis.Med Mycol. 2023 Aug 2;61(8):myad077. doi: 10.1093/mmy/myad077. Med Mycol. 2023. PMID: 37533203 Review.
-
Pathogenicity mechanisms and host response during oral Candida albicans infections.Expert Rev Anti Infect Ther. 2014 Jul;12(7):867-79. doi: 10.1586/14787210.2014.916210. Epub 2014 May 7. Expert Rev Anti Infect Ther. 2014. PMID: 24803204 Review.
References
-
- Nuss AM, Beckstette M, Pimenova M, Schmühl C, Opitz W, Pisano F, Heroven AK, Dersch P. 2017. Tissue dual RNA-seq allows fast discovery of infection-specific functions and riboregulators shaping host-pathogen transcriptomes. Proc Natl Acad Sci U S A 114:E791–E800. doi:10.1073/pnas.1613405114 - DOI - PMC - PubMed
-
- Lemberg C, Martinez de San Vicente K, Fróis-Martins R, Altmeier S, Tran VDT, Mertens S, Amorim-Vaz S, Rai LS, d’Enfert C, Pagni M, Sanglard D, LeibundGut-Landmann S. 2022. Candida albicans commensalism in the oral mucosa is favoured by limited virulence and metabolic adaptation. PLoS Pathog 18:e1010012. doi:10.1371/journal.ppat.1010012 - DOI - PMC - PubMed
-
- López-Agudelo VA, Baena A, Barrera V, Cabarcas F, Alzate JF, Beste DJV, Ríos-Estepa R, Barrera LF. 2022. Dual RNA sequencing of Mycobacterium tuberculosis-infected human splenic macrophages reveals a strain-dependent host-pathogen response to infection. Int J Mol Sci 23:1803. doi:10.3390/ijms23031803 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

