In vivo evolution to hypermucoviscosity and ceftazidime/avibactam resistance in a liver abscess caused by Klebsiella pneumoniae sequence type 512
- PMID: 39171923
- PMCID: PMC11423586
- DOI: 10.1128/msphere.00423-24
In vivo evolution to hypermucoviscosity and ceftazidime/avibactam resistance in a liver abscess caused by Klebsiella pneumoniae sequence type 512
Abstract
Carbapenemase-producing Klebsiella pneumoniae represents a major public health issue globally. Isolates with resistance to the newest drugs, like ceftazidime/avibactam (CZA), are increasingly reported. In this study, we analyzed the evolution of KPC-3-producing sequence type (ST) 512 K. pneumoniae strains isolated at three different times (hospitalization days 45, 56, and 78) from the same patient, two of which were observed in a pericholecystic liver abscess. The three K. pneumoniae isolates (295Kp, 304Kp, and hmv-318Kp) from the same patient were subjected to antimicrobial susceptibility testing, whole-genome sequencing, sedimentation assay, biofilm measurement, serum resistance assay, macrophage phagocytosis, and adhesion assays. KPC-producing isolate hmv-318Kp exhibited carbapenem susceptibility, hypermucoviscous (hmv) colony phenotype and CZA resistance. Virulence markers of hypervirulent Klebsiella were absent. Two non-synonymous mutations were identified in the hmv-318Kp genome comparing with isogenic strains: a single-nucleotide polymorphism (SNP) occurred in the pKpQIL plasmid, changing blaKPC-3 in the blaKPC-31 gene variant, conferring CZA resistance; and a second SNP occurred in the wzc gene of the capsular biosynthesis cluster, encoding a tyrosine kinase, resulting in the F557S Wzc protein mutation. The Klebsiella pneumoniae strain exhibiting an hmv phenotype (hmv-Kp) phenotype has been previously associated with amino acid substitutions occurring in the Wzc tyrosin kinase protein. We observed in vivo evolution of the ST512 strain to CZA resistance and acquisition of hypermucoviscosity. The pathogenetic role of the detected Wzc substitution is not fully elucidated, but other Wzc mutations were previously reported in hmv K. pneumoniae. Wzc mutants may be more frequent than expected and an underreported cause of hypermucoviscosity in K. pneumoniae clinical isolates.
Importance: Here we describe the evolution of KPC-3-producing ST512 K. pneumoniae isolated at three different times from the same patient of which the last one, from a biliary abscess, showed CZA resistance by KPC-31 production and manifested hmv colony phenotype. Hypervirulent Klebsiella pneumoniae (hv-Kp) isolates are increasingly reported worldwide. Their hypervirulent traits are associated with the presence of rmpA/A2 genes and an hmv. In this study, we identified an hmv-Kp that lacked the rmpA-D cluster but showed an amino acid substitution in the Wzc tyrosin kinase protein, involved in the capsular biosynthesis. This hmv-Kp strain emerged in vivo and evolved resistance to ceftazidime/avibactam resistance in a liver abscess of a patient. Our findings suggest that wzc mutations may be underreported, making it challenging to distinguish hv-Kp from "classic" K. pneumoniae with an hmv phenotype.
Keywords: KPC-31; ST512; antibiotic resistance; capsule; hmv; hypermucoid; hypervirulence; mucoviscous; rmpA negative; serum susceptibility; string test; wzc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
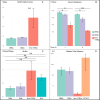

References
-
- Di Pilato V, Errico G, Monaco M, Giani T, Del Grosso M, Antonelli A, David S, Lindh E, Camilli R, Aanensen DM, Rossolini GM, Pantosti A, AR-ISS Laboratory Study Group on carbapenemase-producing Klebsiella pneumoniae . 2021. The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J Antimicrob Chemother 76:355–361. doi: 10.1093/jac/dkaa431 - DOI - PubMed
-
- Taracila MA, Bethel CR, Hujer AM, Papp-Wallace KM, Barnes MD, Rutter JD, VanPelt J, Shurina BA, van den Akker F, Clancy CJ, Nguyen MH, Cheng S, Shields RK, Page RC, Bonomo RA. 2022. Different conformations revealed by NMR underlie resistance to ceftazidime/avibactam and susceptibility to meropenem and imipenem among D179Y variants of KPC β-lactamase. Antimicrob Agents Chemother 66:e02124-21. doi: 10.1128/aac.02124-21 - DOI - PMC - PubMed
-
- Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, Trancassini M, Faino L, Venditti M, Antonelli G, Raponi G. 2021. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 65:e00574-21. doi: 10.1128/AAC.00574-21 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- EU-MUR PNRR PE00000007 PE13 INF-ACT/Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR)
- PRIN 2022 Project 2022FN7ANE/Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR)
- RP12218162DFBF0A and RP1221816196D9A2 and SEED PNR 2021 335/2021/Sapienza Università di Roma (Sapienza)
- Ricerca corrente]" to IRCCS San Raffaele Roma/Ministero della Salute (Italy Ministry of Health)
- Rome Technopole POR Lazio FSE 2014-2020/Sapienza Università di Roma (Sapienza)
LinkOut - more resources
Full Text Sources
Medical
